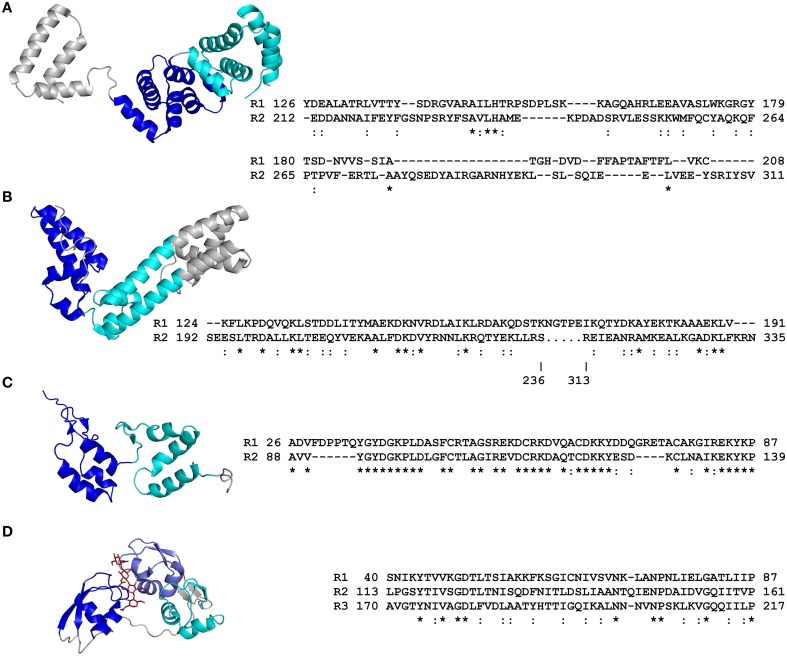
Figure 2.
Primary and tertiary structures of repeat domains from RCP effectors of plant-associated fungi and an oomycete. (A) Crystal structure of the ATR1 effector from the Arabidopsis thaliana oomycete pathogen, Hyaloperonospora arabidopsidis (Protein Data Bank [PDB] code 3RMR; Chou et al., 2011). (B) Crystal structure of the AvrM-A effector from the flax rust fungus, Melampsora lini (4BJN; Ve et al., 2013). (C) Nuclear magnetic resonance (NMR) structure of repeat units 1 and 2 from the candidate effector Cin1 of the apple scab fungus, Venturia inaequalis (2LHT; Mesarich et al., 2012). (D) Crystal structure of the Ecp6 effector from the tomato leaf mold fungus, Cladosporium fulvum. The lysin motif (LysM) repeat units 1 and 3 coordinate the binding of a single chitin tetramer by means of an inter-repeat domain groove (4B8V; Sánchez-Vallet et al., 2013). Structural coordinate files were downloaded from the Research Collaboratory for Structural Bioinformatics (RCSB) PDB (http://www.rcsb.org/pdb/home/home.do). Alternating repeat units are colored blue, slate, and cyan, respectively. Non-repetitive sequence is colored gray. The chitin tetramer in (D) is colored red. An amino acid sequence alignment detailing the primary structure of each RCP effector repeat domain is shown to the right of each tertiary structure (as based on that presented in each tertiary structure). Repeat (R) units are numbered according to their position in the RCP effector. The start and end position of each repeat unit in the full-length RCP effector is shown. Conserved (*) and strongly similar (:) amino acid residues shared between repeat units are shown below the sequence alignment. The figure was prepared using PyMol (https://www.pymol.org/) and Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Structure-based sequence alignments of repeat units from ATR1 and AvrM-A are adapted from Chou et al. (2011) and Ve et al. (2013), respectively.