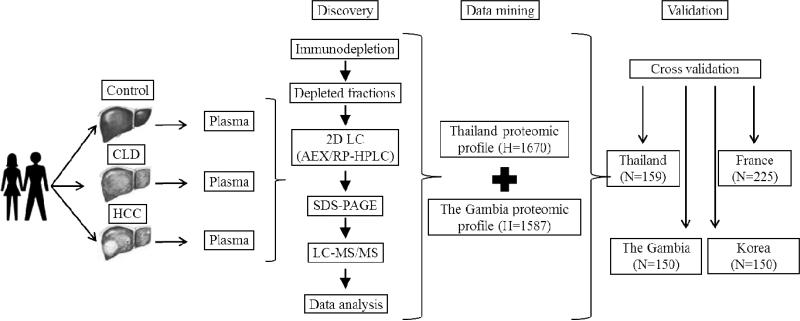
Figure 1. Outline of study design, from protein discovery to biomarker validation.
The study included three steps, with first plasma proteomics, second data mining to identify candidate markers and third validation in several cohorts of patients. First, biospecimens (plasma) were collected in two structured hospital-based case-control studies in two regions of high incidence of HB carriage and of HCC, The Gambia and Thailand, using protocols optimized from plasma proteomics. These case-control studies included control individuals (no liver symptoms), individuals with chronic liver disease patients (CLD, chronic active hepatitis B or hepatitis C), and patients with confirmed diagnosis of HCC. A small number of plasma specimens of cases (HCC, n=10), CLD (n=10) and controls (n=10) from each region were extensively analysed using a deep-plasma proteomic approach. Next, data generated from The Gambia and from Thailand were mined to identify candidate biomarkers that were differentially expressed in cases, CLD and controls in both regions. Third, two of these candidate markers, OPN and LTBP2, were quantified in the entire collection of plasma from the two case-control studies and in two additional series, a case series from France (CLD and/or HCC) and a prospective study of HB carriers from Korea who subsequently developed HCC. H: number of identified proteins; N: number of plasma specimens tested.