Fig. 1.
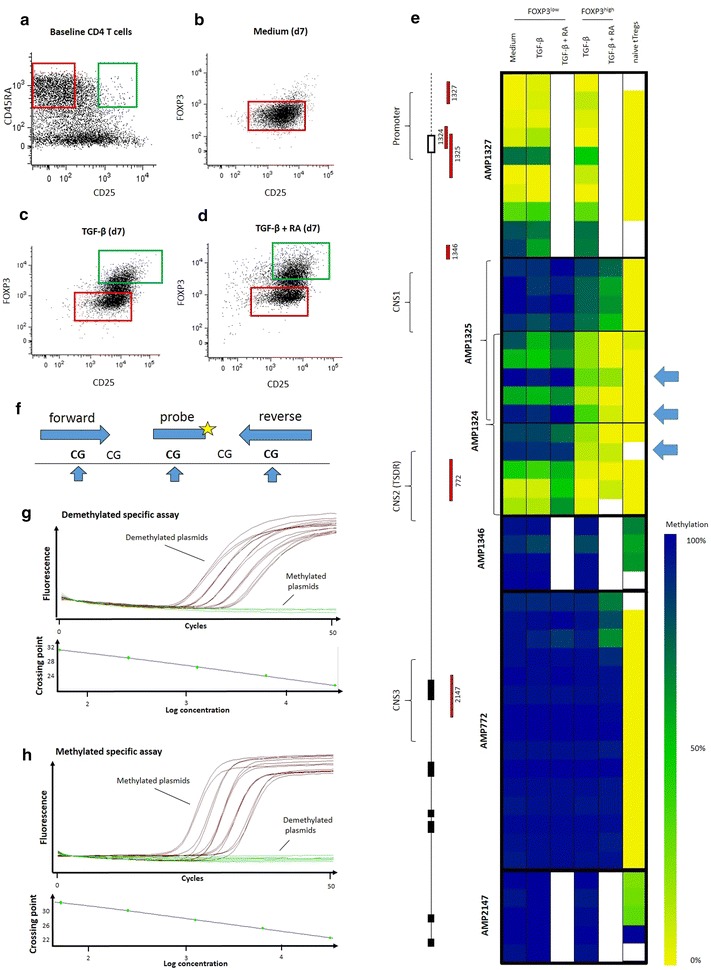
FOXP3 promoter RT-PCR assay design. Naïve T cells (CD4+CD45RA+CD62L+CD25−) were sorted from CD4 enriched buffy coats (a, red gate) and put in culture with IL-2 and CD3/CD28 stimulation in medium alone (b), in the presence of TGF-β (c) or both TGF-β and retinoic acid (d). At day 7, cells were sorted based on FOXP3 expression. FOXP3high (green gates) and FOXP3low samples (red gates), as well as baseline naïve tTreg controls (a, green gate) were pyrosequenced at the promoter and three conserved non-coding sequence regions of the FOXP3 gene (e). Promoter CpG sites at positions −126, −77 and −56 from transcription start site showed the greatest differential methylation in FOXP3high TGF-induced T cells (iTregs) (indicated with arrows). These sites were selected to design primers and RT-PCR probes specific for their methylation status (f). Specificity of methylated and unmethylated RT-PCR primers and probes was validated using unmethylated (g) and methylated (h) plasmid standards dilutions at 31,250, 6250, 1250, 250 and 50 copy numbers per reaction
