Figure 1.
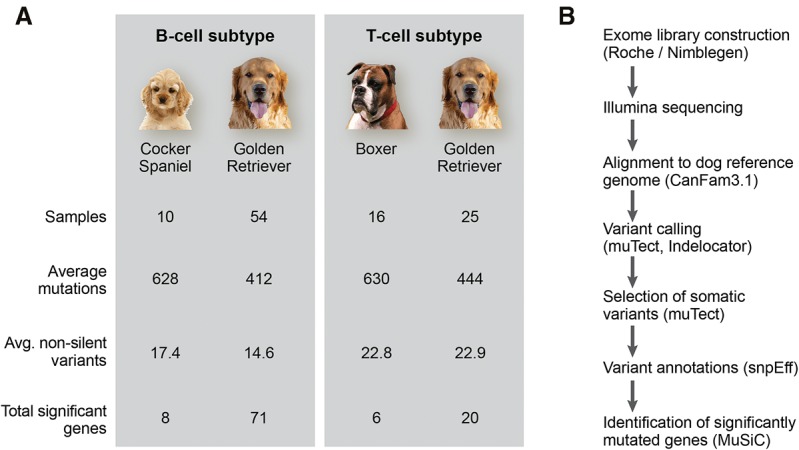
Analysis pipeline and mutation number overview. (A) Sample numbers and average mutations per breed and immunophenotype are indicated. (B) Exome libraries were sequenced and variants were called with MuTect, a caller adapted for cancer data, and IndeLocator. Somatic variants were annotated using SnpEff. MuSiC was used for identification of significantly mutated genes.
