Abstract
In animals, microRNAs frequently form families with related sequences. The functional relevance of miRNA families and the relative contribution of family members to target repression have remained, however, largely unexplored. Here, we used the Caenorhabditis elegans miR-58 miRNA family, composed primarily of the four highly abundant members miR-58.1, miR-80, miR-81, and miR-82, as a model to investigate the redundancy of miRNA family members and their impact on target expression in an in vivo setting. We found that miR-58 family members repress largely overlapping sets of targets in a predominantly additive fashion. Progressive deletions of miR-58 family members lead to cumulative up-regulation of target protein and RNA levels. Phenotypic defects could only be observed in the family quadruple mutant, which also showed the strongest change in target protein levels. Interestingly, although the seed sequences of miR-80 and miR-58.1 differ in a single nucleotide, predicted canonical miR-80 targets were efficiently up-regulated in the mir-58.1 single mutant, indicating functional redundancy of distinct members of this miRNA family. At the aggregate level, target binding leads mainly to mRNA degradation, although we also observed some degree of translational inhibition, particularly in the single miR-58 family mutants. These results provide a framework for understanding how miRNA family members interact to regulate target mRNAs.
MicroRNAs (miRNAs) are noncoding RNA molecules of ∼21 nucleotides in length that have an essential role in post-transcriptional regulation of gene expression in metazoan animals, plants, and protozoa (Fabian et al. 2010). They associate with Argonaute proteins to guide RNA-induced silencing complexes (RISCs) to transcripts in a sequence-dependent manner, thereby down-regulating protein levels of hundreds of different targets (Baek et al. 2008; Selbach et al. 2008; Bartel 2009). The current version of miRBase (Kozomara and Griffiths-Jones 2014) catalogs 1872 human miRNAs and 223 Caenorhabditis elegans miRNAs. Due to their broad repertoire of targets, miRNAs are predicted to control the activity of >60% of all protein-coding genes and to be involved in almost every cellular process described to date (Bushati and Cohen 2007; Ghildiyal and Zamore 2009). Misregulation of miRNA expression has been associated with numerous diseases (Calin and Croce 2006; Nana-Sinkam and Croce 2011; Mendell and Olson 2012; Maciotta et al. 2013).
Many lines of evidence indicate that the “seed” region of the miRNA, which spans the 6–8 nt at the 5′ end of the miRNA, mediates the interaction with the mRNA target (Bartel 2009). Targets that are perfectly complementary to the miRNA seed are known as canonical. Complementary sequences outside of the seed region can further stabilize the interaction between miRNA and its target (Grimson et al. 2007). Noncanonical targets have also been reported, although in much smaller numbers (Lal et al. 2009; Shin et al. 2010; Chi et al. 2012; Loeb et al. 2012; Helwak et al. 2013; Grosswendt et al. 2014). Different studies have reported various degrees of miRNA-induced mRNA destabilization and translational inhibition (Baek et al. 2008; Selbach et al. 2008; Hendrickson et al. 2009; Guo et al. 2010; Huntzinger and Izaurralde 2011; Jovanovic et al. 2012; Stadler et al. 2012; Hausser et al. 2013). A few recent studies have shown that translational inhibition precedes mRNA destabilization (Bazzini et al. 2012; Djuranovic et al. 2012), although a report argued that miRNA-induced poly(A)-tail shortening can affect translational repression and mRNA destabilization differently in different developmental stages (Subtelny et al. 2014). Thus, in spite of the intensive research on the mechanisms of miRNA target regulation over the last decade, much remains uncharted.
Based on the seed sequence similarity, miRNAs are grouped into families (Lewis et al. 2005). Sixty-five percent of all miRNAs are part of multimember families (Wang et al. 2014). Many of the miRNA families in humans regulate targets relevant for development and disease. For example, the cluster of miR-17-92 miRNAs, containing four distinct seed families, is oncogenic, promotes proliferation, inhibits differentiation, and increases angiogenesis (Concepcion et al. 2012), whereas miR-29 family members (miR-29a-c) act to induce apoptosis and have been found to be down-regulated in various cancers (Iorio et al. 2005; Pekarsky et al. 2006; Yanaihara et al. 2006; Porkka et al. 2007; Park et al. 2009). Large-scale studies of miRNA deletions in C. elegans revealed strong phenotypes in several family mutants, while most single miRNA deletions had no effect on development or viability (Miska et al. 2007; Alvarez-Saavedra and Horvitz 2010; Shaw et al. 2010). At least in some cases, family members likely function redundantly, since expression of any miRNA family member can rescue the family mutant phenotypes (Alvarez-Saavedra and Horvitz 2010). However the actual extent of the miRNA family member redundancy in humans and model organisms, the extent of overlap in their targets, and their relative contribution in target repression are still poorly understood.
In this study we used the C. elegans miR-58 miRNA family as a model to investigate target regulation by a miRNA family in a living organism. The miR-58 family is by far the most abundant family in C. elegans, accounting for >30% of all total miRNAs (Jan et al. 2011). It is composed of four highly abundant members—miR-58.1, miR-80, miR-81, and miR-82—and two low abundance miRNAs—miR-58.2 and miR-2209.1—that were more recently discovered. Single-deletion mutants of the miR-58 family do not display obvious abnormalities, but a mir-80; mir-58.1; mir-81-82 quadruple mutant shows defects in locomotion, body size, egg laying, and dauer formation (Alvarez-Saavedra and Horvitz 2010). Here we show that miR-58 family members regulate their targets in a primarily additive fashion, with target level threshold critical for the phenotype largely influenced by all the family members. Interestingly, miR-58 family members do not compensate each other's expression, indicating a lack of feedback mechanism between these family members. Furthermore, we describe the molecular consequences on target RNA and protein expression levels following the dismantling of the most abundant C. elegans miRNA family, which indicate that miR-58 family members exert both translation inhibition and mRNA degradation on their targets, with eventual mRNA degradation being the dominant consequence of miRNA repression.
Results
Proteomic consequences of miR-58 family member mutations reveal cumulative nature of miR-58 target repression
To investigate the contribution of individual miR-58 family members on target mRNA and protein abundance, we undertook a combined proteomic and transcriptomic analysis of the various single and multiple miR-58 mutant backgrounds available in C. elegans.
We utilized selected reaction monitoring (SRM) assays to quantify 22 proteins whose corresponding transcripts are predicted by TargetScan (Friedman et al. 2009; Jan et al. 2011) to be miR-58 family targets, as well as 22 random control proteins (Jovanovic et al. 2012), in all possible combinations of the three available miR-58 member deletions: mir-80, mir-58.1, and mir-81-82 (Supplemental Fig. S1). For a comprehensive analysis of miR-58 family impact on the proteome, we further used a shotgun proteomic approach (in two variants: with and without hydrophilic interaction liquid chromatography [HILIC] fractionation) to characterize the proteome of mir-58.1 single mutants, mir-80; mir-58.1 double mutants, and mir-80; mir-58.1; mir-81-82 quadruple mutants (Supplemental Fig. S2A). Unfractionated SILAC measurement quantified 644 proteins in the wild type, 657 proteins in the mir-58.1 single mutant, 661 proteins in the mir-80; mir-58.1 double mutant, and 663 proteins in the mir-80; mir-58.1; mir-81-82 quadruple mutant, whereas SILAC with HILIC fractionation quantified 2127, 2236, 2405, and 2396 proteins in the respective samples. The correlation between the intensities of light-labeled peptides was used to measure the reproducibility between different SILAC experiments. Comparison of three unfractionated SILAC replicates between each other and of unfractionated and fractionated SILAC showed very strong and positive correlation (r ≥ 0.9 between unfractionated replicates and r ≥ 0.77 for fractionated vs. unfractionated) (Supplemental Fig. S3). Five hundred eighty-seven and 1781 proteins could be quantified in all four samples with the two distinct SILAC methods, enabling us to analyze system-wide perturbations induced by miRNA family member mutations.
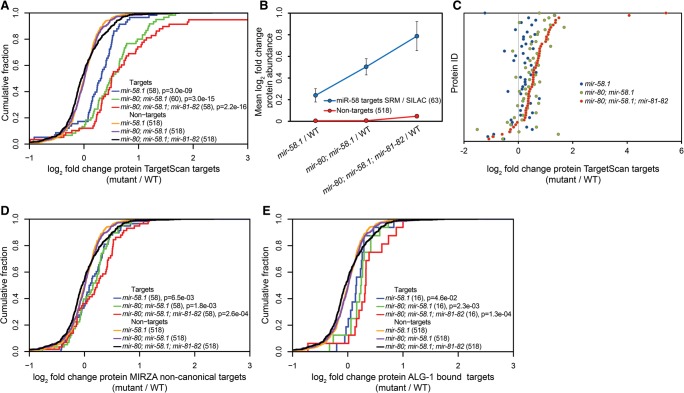
Together, the two mass spectrometric approaches allowed us to quantify the abundance of 63 proteins predicted to be miR-58 family targets by TargetScan (henceforth simply called targets) (Supplemental Fig. S2B), and to compare them to a control group of all 518 nontargets for which we could quantify both the protein and mRNA abundance (see below) (Fig. 1A). As shown in Figure 1B, the effect of miR-58 family members on their targets is cumulative: The increase in target protein abundance compared with the nontargets increases with the number of miRNAs that are deleted. Strikingly, even though at the phenotypic level the single and double mutants are similar to the wild type (Alvarez-Saavedra and Horvitz 2010), the miR-58 targets are significantly up-regulated in these mutants compared with the wild type (mir-58.1, P = 3 × 10−9 and mir-80; mir-58.1, P = 3 × 10−15, Kolmogorov-Smirnov [KS] test) (Fig. 1A). As has been described in other situations, the extent of target up-regulation in average is relatively weak even when all miR-58 family members are deleted (less than twofold) (Fig. 1B,C). Whether the defects observed in the quadruple mutant result from the cumulative effect of these many small changes or whether they result from more significant changes in individual or small numbers of targets (some of which we detected, others we certainly missed, either because the change is tissue specific and restricted to a limited number of cells, or because the protein was not quantified in our study) remains to be determined.
Figure 1.
A combined selected reaction monitoring (SRM) and SILAC approach reveals additive target protein regulation of miR-58 family members. (A) Cumulative distributions of log2 fold expression changes of the 63 TargetScan-predicted miR-58 targets in the mir-58.1, mir-80; mir-58.1, and mir-80; mir-58.1; mir-81-82 mutants relative to the wild type (WT) indicate increased protein abundance with each miR-58 family mutation introduced. Quantified TargetScan-predicted targets identified in SRM, fractionated SILAC, and unfractionated SILAC were compared to a group of 518 nontargets identified in unfractionated SILAC measurements for which we also had transcript abundance data. P-values were calculated using Kolmogorov–Smirnov (KS) test comparing the fold change distributions (log2) of targets and nontargets. (B) Mean log2 fold changes of protein abundances of miRNA targets and nontargets in the different miR-58 family mutants relative to the WT indicate a cumulative effect of miRNA mutations. (C) Differential expression of 55 TargetScan-predicted miR-58 family targets quantified by SRM and unfractionated and fractionated SILAC in mir-58.1, mir-80; mir-58.1, and mir-80; mir-58.1; mir-81-82 relative to the WT. The protein ID indicates the specific protein quantified in different mutants relative to the WT. Proteins are sorted according to increasing up-regulation in the mir-80; mir-58.1; mir-81-82 quadruple mutant. It is apparent that each additional miRNA mutations further increases the expression of TargetScan-predicted targets compared with random controls. (D) Cumulative distributions of log2 protein-level fold changes of 58 noncanonical targets identified by MIRZA (score cutoff > 10) quantified by unfractionated and fractionated SILAC and of 518 nontargets described above in different miR-58 family mutant backgrounds. P-values were calculated using KS test comparing the fold change distributions (log2) of targets and nontargets. (E) Cumulative distributions of log2 protein-level fold changes of ALG-1–bound miR-58 family targets and of 518 nontargets described above in different miR-58 family mutant backgrounds compared with the WT. Requiring that a target reported by Grosswendt et al. (2014) is supported by at least five reads yielded 43 miR-58 family targets; of these, we obtained SILAC data for 16 targets. P-values were calculated using KS test comparing the fold change distributions (log2) of targets and nontargets.
As TargetScan only predicts canonical targets, we applied the MIRZA biophysical miRNA–mRNA interaction model (Khorshid et al. 2013) to predict additional, noncanonical target sites for the miR-58 family miRNAs. At a MIRZA score cutoff of 10, we obtained 58 noncanonical candidate targets that were quantified in our SILAC experiment (Supplemental Table 1). Interestingly, these candidate targets were also significantly up-regulated as a group in mir-58.1 single, mir-80; mir-58.1 double, and mir-80; mir-58.1; mir-81-82 quadruple mutants, albeit to a much lesser extent than the canonical targets predicted by TargetScan (Fig. 1D).
miRNA target prediction programs take into account many factors that probably contribute to miRNA-target interaction such as the extent of seed complementarity, the free energy of miRNA:mRNA complex, the target site accessibility and relative location in the 3′ UTR, as well as the expression of target transcript in the tissue of interest. What is much more difficult to account for is the cellular context, where many RNA-binding proteins are competing for, and possibly masking, the miRNA-binding sites. For this reason, biochemical approaches have been developed to identify miRNA target sites. Recently, an Argonaute ALG-1 photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) protocol was enhanced with a ligation step that enabled the investigators to experimentally detect 3627 miRNA:target conjugates in C. elegans (Grosswendt et al. 2014). We extracted the miR-58 family conjugates from the respective study and investigated how these ALG-1–bound miR-58 family targets behaved in our proteomics data set. Requiring that the miRNA-target conjugates are robustly supported by more than five reads yielded 43 targets, out of which we had quantitative data for 16 (Fig. 1E). Although this is a relatively small number, the response of these 16 targets was similar to that of the predicted targets, with the degree of derepression increasing with the number of mutated miR-58 family miRNAs. We therefore conclude that, at least for the targets that we identified through proteomics, the miR-58 family members appear to have largely additive effects on their targets.
Additive shifts in the mRNA abundance of miR-58 targets with progressive miR-58 family mutations
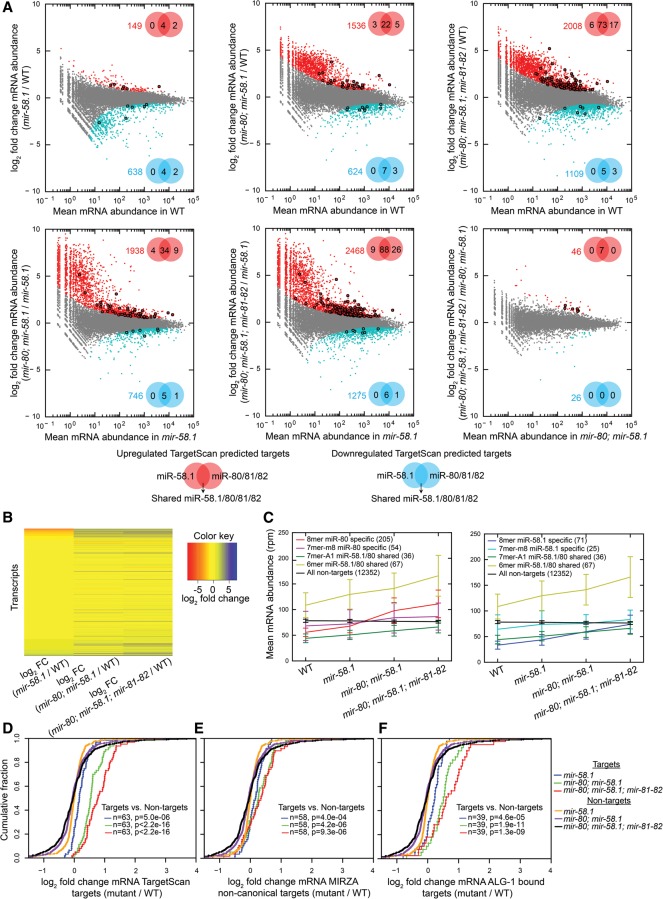
In order to obtain an exhaustive view of the action of miR-58 family members on the RNA level of their targets, we analyzed the transcriptome in different miR-58 family mutants. The reproducibility between three biological replicates was very high (Pearson's product moment correlation ≥0.89) (Supplemental Fig. S5). Because the miR-58.1 and the miR-80/81/82 group of miRNAs differ in a single nucleotide in the seed sequence, TargetScan separates target predictions for these two subfamilies, although their target overlap is ∼80%. We found only minor changes in target mRNA abundance in the mir-58.1 single mutant with six significantly up-regulated [P(adjusted) < 0.01] TargetScan-predicted miR-58.1/80/81/82 targets (Fig. 2A). The target mRNA abundances are more significantly affected in the mir-80; mir-58.1 double mutants and in the mir-80; mir-58.1; mir-81-82 quadruple mutant, with a significant up-regulation of 30 and 96 miR-58 family targets, respectively. The global changes in mRNA abundance, including changes of miR-58 family targets and a much larger number of secondary changes in transcripts that are not predicted to be direct targets of these family members, also become more prominent with increasing depletion of miR-58 family members, reaching 2008 significantly up-regulated and 1109 significantly down-regulated transcripts in the quadruple mutant (Fig. 2A,B). However, the magnitude of changes of miRNA targets is modest and comparable to the magnitude of the protein level changes that we observed in SILAC/SRM experiments (Fig. 2D). The relatively modest mRNA level differences between the double and the quadruple mutants (Fig. 2A, bottom right panel) are in striking contrast with the pronounced differences observed between these two strains at the phenotype level.
Figure 2.
Mutations in individual miR-58 family members additively increase target RNA levels. (A) Plots of the log2 fold change in mRNA abundance between the conditions indicated on the y-axes against the expression level in the condition indicated on the x-axes. Transcripts deemed by DESeq (Anders and Huber 2010) significantly up-regulated or down-regulated transcripts with P(adjusted) < 0.01 are shown as red and blue dots, respectively, and their number is indicated in the upper right or lower right part of the plot. Significantly up-regulated (red) and down-regulated (blue) TargetScan-predicted miR-58.1, miR-80/81/82, and shared miR-58.1/80/81/82 targets are marked with bold dots, and their numbers are indicated in the Venn diagrams. (B) Heatmap indicating differential mRNA expression levels (log2 fold change [FC]) of 16,309 transcripts across different miR-58 family mutants. (C) Average mRNA abundances (in reads per million [rpm]) of groups of transcripts identified in all four conditions containing different seed matches—8mer, 7mer-m8, 7mer-A1, 6mer—and without seed match (in wild type, mir-58.1, mir-80; mir-58.1, and mir-80; mir-58.1; mir-81-82) indicate average contributions of miR-58 family members in their target up-regulation. TargetScan-predicted miR-58.1– and miR-80/81/82–specific and overlapping targets show similar additive trends. Error bars, SD. (D–F) Cumulative distributions of log2 fold changes of the following and 518 nontargets in different miR-58 family mutants compared with WT: (D) 63 TargetScan-predicted targets, (E) 58 noncanonical targets identified by MIRZA (score cutoff > 10), and (F) 39 ALG-1–bound miR-58 family targets (Grosswendt et al. 2014). P-values were calculated using a KS test comparing the fold change distributions (log2) of targets and nontargets.
We categorized the 595 TargetScan-predicted targets that were quantified at mRNA level into seven groups based on fold changes relative to the wild type (Supplemental Table 2). As previously found at the protein level, the largest group of responders (93 targets) showed additive up-regulation with increasing loss of miR-58 family members. A smaller group of 19 targets exhibited changes that could be indicative of synergistic regulation. Three groups contained targets shared between two or three, but not all, miR-58 family members. Finally, 53 targets appeared to be family member specific, as they responded only to deletion of that one member. The last group contained all 270 nonresponsive targets (fold change in mir-80; mir-58.1; mir-81-82 quadruple mutant < 1.3), which are likely to be false predictions or might be targeted at a different developmental stage.
The extent of target abundance change was to some extent dependent on the predicted strength of interaction between miRNAs and targets. When 8mer, 7mer-m8, 7mer-A1, and 6mer (Bartel 2009) were compared with nontargets, the P-value was in general decreasing with the increased target site strength (Supplemental Fig. S6). However, many of the mRNAs that were predicted by TargetScan to respond to only one of the two subfamilies did in fact respond to the mutation of miRNAs from both families (Fig. 2C). Conversely, there was no enrichment in TargetScan-predicted miR-58.1– and miR-80-81-82–specific targets in the group of experimentally observed specific targets (Supplemental Table 2). This suggests that in vivo, strict complementarity to the miRNA seed is not absolutely required for target recognition. Our conclusion is consistent with previous observations; for example, C. elegans let-7 is known to regulate lin-41 via imperfect base-pairing (Vella et al. 2004a,b).
We also studied the changes in mRNA abundance of the noncanonical targets that we previously predicted using MIRZA and of the targets identified in the ALG-1–bound data set (Grosswendt et al. 2014). Noncanonical targets identified by MIRZA showed significant increases in mRNA abundance in the mir-58.1 single, mir-80; mir-58.1 double, and mir-80; mir-58.1; mir-81-82 quadruple mutants (Fig. 2E). The additive effect of miR-58 family mutations on the RNA level was also observed on the 39 quantified targets out of the 43 obtained from the ALG-1 data set (Fig. 2F). Taken together, our data show that miR-58 family members widely cooperate, in the sense of working additively, to down-regulate target mRNA and protein levels.
Although the predicted miR-58 TargetScan targets had increased target mRNA and protein levels in different miR-58 family mutants, it is still possible that this is a consequence of secondary effects. To address this issue, we selected seven highly up-regulated targets quantified in SRM, fractionated SILAC, unfractionated SILAC, and RNA-seq (C30B5.7, C37H5.13, cgh-1, isw-1, lys-1, clec-89, and nas-3) for further validation with dual-luciferase reporters in HeLa cells, a heterologous assay that was previously used to examine C. elegans target repression (Lytle et al. 2007). Six out of the seven (85%) tested targets (all but lys-1) were repressed following miR-58.1 or miR-80 transfection in a site-specific manner, suggesting that indeed they are repressed by miR-58 family in vivo and that the miR-58 target site is necessary for repression (Supplemental Fig. S7). This conclusion is further supported by the recent demonstration that the pmk-2 p38 MAPK, for which we had detected 1.5-, 2.3-, and 4.8-fold up-regulation in mir-58.1 single, mir-80; mir-58.1 double, and mir-80; mir-58.1; mir-81-82 quadruple mutant samples, is a bona fide miR-58 family target (Pagano et al. 2015).
Reduced translational efficiency of derepressed targets
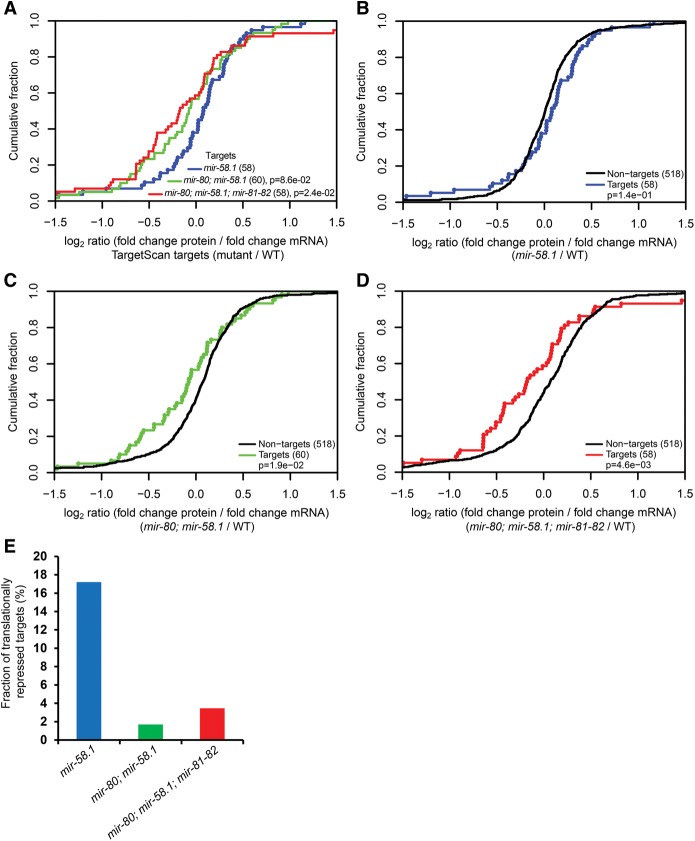
Having measured protein and mRNA abundances of 63 different targets in various miR-58 family mutants, we further sought to determine the relative target mRNA destabilization and translation repression that are induced by subgroups of miR-58 family miRNAs in the context of the organism. As previously reported, we found a slight tendency for the increase in protein abundance to be greater than the increase in mRNA abundance in the mir-58.1 mutant (Fig. 3A). For example, SRM data on 22 verified targets (Supplemental Fig. S8, upper row; Jovanovic et al. 2012) showed modest target protein abundance shifts in single miR-58 family mutants, which reached significance in the mir-58.1 single mutant (P = 0.012, KS test), whereas the RNA levels of these targets were found unchanged relative to random controls by qRT-PCR (Supplemental Fig. S9, upper row). Furthermore, the proportion of the miR-58 family targets that were up-regulated on the protein level with no change in RNA levels was 17.2% in the mir-58.1 single mutant, compared with only 1.7% and 3.5% in the mir-80; mir-58.1 double and mir-80; mir-58.1; mir-81-82 quadruple mutants, respectively (Fig. 3E). Finally, the correlation between target protein and mRNA levels was significantly lower in mir-58.1 single mutants than in the double and quadruple mutants (Supplemental Fig. S10).
Figure 3.
Reduced relative translation efficiency of miR-58 targets in multiple, but not single, miR-58 family mutants. (A–D) Translational efficiency calculated as log2 (fold change protein abundance/fold change mRNA abundance) for targets and 518 nontargets. P-values were calculated using a KS test comparing the fold change distributions (log2) of targets in mir-80; mir-58.1 double and mir-80; mir-58.1; mir-81-82 quadruple mutants to the mir-58.1 single mutant (A) and of targets to nontargets (B–D). (E) Release of translational inhibition is particularly prominent in the single mir-58 mutant. Fraction of translationally repressed [log2 fold change protein abundance (mutant/WT) > 0.05, log2 mRNA abundance (mutant/WT) < 0.05] TargetScan-predicted targets in mir-58.1 single, mir-80; mir-58.1 double, and mir-80; mir-58.1; mir-81-82 quadruple mutants.
In contrast, mir-80; mir-58.1 double and mir-80; mir-58.1; mir-81-82 quadruple mutants showed a significantly stronger increase in mRNA abundance than in protein abundance (Fig. 3C,D; Supplemental Figs. S8, S9). This indicates, surprisingly, a lower translation efficiency in mutants with little to no miR-58 family activity. As distinct tissues and cellular contexts can employ different mechanisms for miRNA silencing (Subtelny et al. 2014) or use different 3′ UTR isoforms (Blazie et al. 2015), we analyzed whether miR-58 targets expressed in the germline or in the soma might be regulated differently. Even though we observed similar additive effects on protein and target mRNA regulation, we found that the observed reduced translational efficiency in the miR-58 family mutants stems from the germline-expressed targets (Supplemental Fig. S11). This finding further substantiates the importance of studying different cellular contexts separately when exploring miRNA-mediated target repression.
miR-58 family members fail to sense each other
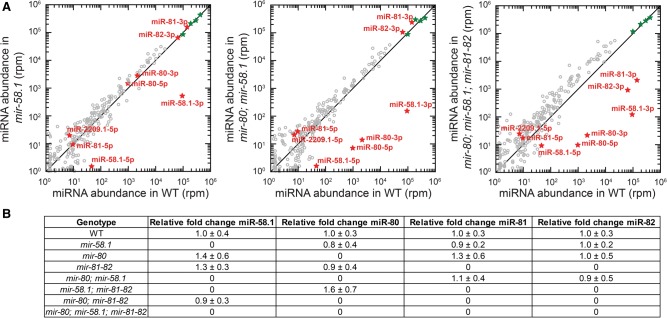
We next investigated whether miR-58 family members compensate each other's expression. Small RNA profiling revealed that the miR-58 family is by far the most abundant group of miRNAs in the fourth larval stage of C. elegans, with miR-58.1, miR-81, and miR-82 representing 9%, 14%, and 6%, respectively, of the miRNA population. miR-80 has a lower expression, corresponding to 0.3% of the miRNA population, whereas the two recently discovered members of the miR-58 family, miR-2209.1 and miR-58.2, are essentially undetectable (two and zero reads per million, respectively). The correlation between the three replicates of miR-58 family mutants was high (Pearson's product moment correlation r ≥ 0.75) (Supplemental Fig. S12).
Neither small RNA profiling nor miRNA qRT-PCR identified significant changes in the expression of miRNAs other than those that were deleted in the mutant backgrounds (Fig. 4A,B). These results suggest that miR-58 family members do not sense each other's expression and do not compensate for the loss of expression of other family members. These results are consistent with the largely additive effect observed at the target level with progressive loss of family members. Because of the high abundance of the miR-58 family, the removal of all major family members (∼30% of all miRNAs at the fourth larval stage) is expected to significantly increase the availability of Argonaute molecules for interaction with other miRNAs. This may stabilize these miRNAs, consistent with the modest general increase in the abundance of all other miRNAs that we observed in the mir-80; mir-58.1; mir-81-82 quadruple mutants. This effect is likely further enhanced by the fact that ALG-1, one of the main two miRNA Argonautes in C. elegans, is itself a target of the miR-58 family. Indeed, MIRZA analysis identified ALG-1 as one of the best miR-58 family candidate targets, with both canonical (MIRZA score, 50.74) and noncanonical target sites (MIRZA score, 51.56) (Supplemental Table 1).
Figure 4.
Loss of miR-58 family members does not lead to any compensation effects among other family members. (A) Small RNA sequencing comparing miRNA abundance in reads per million between different miR-58 family mutant samples and the WT. The reads are normalized to the four calibration sequences that were added in the sample in equal amounts during miRNA library preparation (marked as green stars on the plot). (B) miRNA-qRT-PCR assaying relative fold change of miR-58.1, miR-80, miR-81, and miR-82 in different miR-58 family mutant backgrounds identified no significant changes. The levels were normalized to the miRNAs stably expressed throughout the development, miR-250 and miR-52 (Kato et al. 2009), and are relative to WT. Standard deviations between three biological replicates are indicated.
Loss of miR-58 family impairs IR-induced germ cell apoptosis
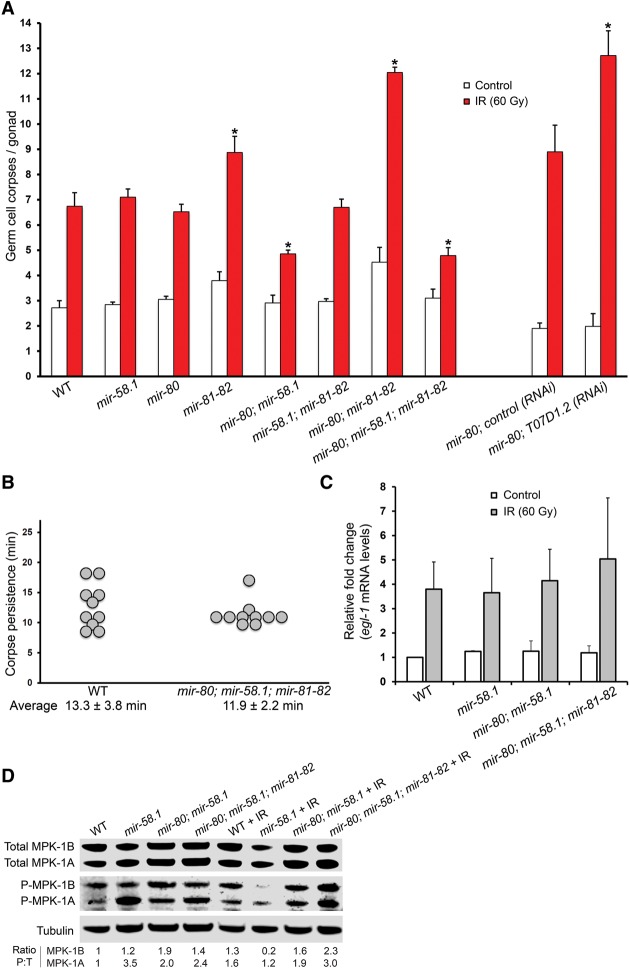
The miR-58 family miRNAs of C. elegans are homologous to the bantam miRNA of Drosophila, which has been previously found to inhibit apoptosis by targeting the proapoptotic factor hid (Brennecke et al. 2003). Moreover, the gene ontology (GO) term analysis of 509 genes up-regulated in the quadruple mutant on the protein level (log2 fold change (mir-80; mir-58.1; mir-81-82/WT) > 0.3, fractionated SILAC) was weakly enriched in the term “cell death” (P = 0.04), associated with the up-regulation of nine proteins (CAR-1, CGH-1, CLP-1, CLP-7, DRP-1, GFI-1, LAM-1, M57.2, and NUC-1). These observations made us explore the potential role of miR-58 family members in the regulation of apoptosis. We found that both the mir-80; mir-58.1 double mutant and the mir-80; mir-58.1; mir-81-82 quadruple mutant exhibit a reduced response in ionizing radiation (IR)–induced apoptosis, whereas the mir-81-82 double mutant and the mir-80; mir-81-82 triple mutant showed an increase in IR-induced germ cell apoptosis (Fig. 5A). However, because the 6-kb deletion of mir-81-82 covers the T07D1.2 gene, whose RNAi resulted in increased levels of DNA damage–induced apoptosis (Fig. 5A), it is likely that the increase of germ cell corpses in the mir-81-82 background occurred due to the deletion of T07D1.2 and not due to different apoptosis-related target specificity of miR-81 and miR-82 compared with other miR-58 family members. Reintroduction of either miR-58.1 or miR-80 from a transgene could partially restore the IR-induced apoptosis of the miR-58 family mutant, confirming that the phenotype of the quadruple mutant is likely caused by the lack of miR-58 family members and not by other differences in the genetic background (Supplemental Fig. S13).
Figure 5.
miR-58 family mutants show defective apoptotic response following irradiation. (A) mir-80; mir-58.1 and mir-80; mir-58.1; mir-81-82 mutants show decreased germline apoptosis following IR, while mir-81-82 double and mir-80; mir-81-82 triple mutant show an increase in IR-induced apoptosis levels. RNAi of T07D1.2, a gene largely deleted in nDf54 deficiency that covers miR-81 and miR-82 locus, induces germline apoptosis following IR. Empty vector was used as control RNAi. Synchronized animals in L4 larval stage were irradiated, and DNA damage–induced germline apoptosis was quantified 24 h after irradiation. Error bars, SEM from three biological replicates (20 worms were scored per experiment); asterisk, P-value for a paired t-test comparing the mutants/T07D1.2 RNAi to the WT/empty vector control (*P < 0.001 IR samples). (B) Apoptotic clearance is not impaired in the mir-80; mir-58.1; mir-81-82 quadruple mutant. Time of corpse persistence was assayed by measuring the time from the corpse's first appearance until it was no longer visible. Average persistence and SD are denoted below the chart. (C) Transcriptional up-regulation of egl-1 following IR is normal in the various miR-58 family mutants. mRNA was extracted 12 h after irradiation of L4 larvae, and egl-1 levels were assayed by quantitative real-time PCR. mRNA levels for each sample were normalized to three housekeeping genes—pgk-1, cdc-42, and Y45F10—and the fold induction was calculated relative to wild-type untreated worms. Error bars, SEM (n = 2). (D) Levels of MPK-1 activation (phosphorylation) differ between various miR-58 family mutants. Protein was extracted from irradiated and nonirradiated young adult worms, and equal amounts were loaded onto SDS-PAGE gels. Total MPK-1 was detected with an anti-ERK-antibody, activated MPK-1 was detected with an anti-P-ERK antibody, and a-tubulin was used as a loading control. The ratio of activated MPK-1 (P) to total MPK-1 (T) normalized against wild-type nonirradiated worms is shown for the MPK-1A and MPK-1B isoforms.
Reduced germ cell corpse numbers following IR can be caused by defects in cell death activation or through faster clearance of these corpses following their death. To determine whether engulfment kinetics were affected, we measured corpse persistence in the mir-80; mir-58.1; mir-81-82 quadruple mutant and found no difference relative to the wild type (Fig. 5B). Thus, reduced apoptosis levels are the likely cause of the observed phenotype.
Because developmental cell death and physiological germ cell deaths are normal in the mutants (Fig. 5A; Supplemental Fig. S14), we surmised that loss of miR-58 family affects DNA damage response upstream of the core apoptotic machinery. In C. elegans, DNA damage is sensed by the TP53 homolog CEP-1, which induces transcriptional activation of the BH3-only proapoptotic genes egl-1 and ced-13 (Schumacher et al. 2001, 2005). EGL-1 and CED-13 in turn trigger the downstream apoptotic response (Conradt and Horvitz 1998; Schumacher et al. 2001). Since the mir-80; mir-58.1; mir-81-82 quadruple mutant showed a reduced DNA damage–induced germ cell apoptosis response, we tested if CEP-1 activation following irradiation was altered in these mutants by essaying egl-1 induction with quantitative RT-PCR (Fig. 5C). We found no difference in egl-1 induction compared with the wild type, suggesting that loss of miR-58 family influences apoptosis downstream from or in parallel to CEP-1.
Several recent studies identified MPK-1 as a central regulator of germ cell death (Rutkowski et al. 2011; Eberhard et al. 2013). We therefore investigated whether the reduced apoptotic response following DNA damage may be due to a reduction in MPK-1 phosphorylation. MPK-1 is expressed in two isoforms: MPK-1B, predominantly expressed in the germline, and MPK-1A, predominantly expressed in the soma (Lee et al. 2007). Interestingly, the mir-80; mir-58.1; mir-81-82 quadruple mutant does not show strong changes of MPK-1B induction following ionizing irradiation. In contrast, the mir-58.1 single mutant shows a very strong and the mir-80; mir-58.1 double mutant a slight decrease in MPK-1B isoform activation (Fig. 5D). The absence of direct correlation between impaired MPK-1 activation and defective apoptosis suggests that the modulation of apoptosis by the miR-58 family is complex, involving pathways beyond MPK-1. Indeed, whether the apoptosis defect is caused by the up-regulation of a miR-58 family target or whether it is a secondary side-effect of miR-58 family loss needs to be further investigated.
Discussion
miR-58 family members regulate targets additively
In this study, we explored the relative contribution of miRNA family members to their target repression by taking advantage of an extensive set of deletion mutants for the miR-58 family in C. elegans. Our results suggest that miR-58 family members act mostly additively to down-regulate a largely overlapping set of targets without sensing each other's expression levels. The changes in protein and RNA abundance of miR-58 family targets in various miR-58 family mutants are generally modest (less than twofold for the majority of targets). Subtle changes of protein and RNA abundance following miRNA transfection or in a knock-down study have been reported previously (Baek et al. 2008; Selbach et al. 2008; Yang et al. 2010). However, here we describe the cumulative nature of target regulation by a miRNA family in the setting of a whole organism, leading to much stronger expression changes in the multiple miRNA mutants compared with the single mutants. Consistent with our observations, a recent study illustrated additive mRNA destabilization of pmk-2 in non-neuronal tissues by the miR-58 family members (Pagano et al. 2015). The increasing effects on direct targets were accompanied by an increasing number of secondary perturbations, reaching a significant fraction of the transcriptome and proteome in the quadruple mutant. While mRNA and protein abundance changes are detectable already in the single miR-58 family mutants, locomotion, body size, egg laying, and dauer formation defects have only been reported in the quadruple mutant, in which all four abundant family members are deleted (Alvarez-Saavedra and Horvitz 2010). Detailed phenotypic analyses are likely to identify more subtle defects also in the other mutants. Indeed, we observed, for example, a slight developmental delay in mir-80; mir-58.1 double and mir-58.1; mir-81-82 triple mutants, suggesting that different phenotypes may have different target thresholds.
miR-58 family members have largely overlapping target sets
To what extent the miRNA family members have overlapping target sets is still a matter of intensive discussion (Gregory et al. 2008; Park et al. 2008; Kim et al. 2013). Interestingly, in the setting of the whole organism, miR-58 family members appear to be quite promiscuous, and many targets predicted by TargetScan to be miR-58.1 specific are also efficiently down-regulated by miR-80/81/82 members, whose seed sequence differs at position 8 from the miR-58.1 seed sequence. A possible explanation could be that the position 8 is less important for target selection compared with other positions in the miRNA seed.
A recent study reported various degrees of overlap in the targets bound by different members of miRNA families in the context of ALG-1 (Grosswendt et al. 2014). By analyzing the expression changes of the targets inferred in the Grosswendt study in the miR-58 family mutants, we observed a predominantly additive behavior even on several targets containing noncanonical sites. The observed additive effect among noncanonical targets is likely a consequence of miR-58 family members binding different sites of a given transcript. Of note, as described before in mammalian cell lines (Khorshid et al. 2013), the extent of up-regulation observed among noncanonical targets in vivo was smaller compared with the canonical targets, emphasizing that efficient down-regulation of miRNA targets requires full seed sequence complementarity.
In addition to common targets, our data as well revealed several dozen targets that respond specifically to one miR-58 family member. Based on transcriptional reporters of miRNA promoter activity, miR-58.1, miR-80, miR-81, and miR-82 are coexpressed in some tissues but also show tissue specificities in others (Liu et al. 2008; Martinez et al. 2008; Isik et al. 2010; Kudlow et al. 2012). Thus, target specificity may originate from a miR-58 family member–specific pairing outside of the seed region or from the tissue-specific expression of miR-58 family members.
miR-58 family members destabilize target mRNA and repress its translation
Previously we reported that loss of miR-58.1 resulted in stronger up-regulation at the protein than at the mRNA level, suggesting that translational repression contributes significantly to target regulation by this miRNA (Jovanovic et al. 2012). In our expanded study of all miR-58 family members, we again observed a slight tendency for the protein changes to be greater than the RNA changes in the single miRNA mutants (Fig. 3; Supplemental Figs. S8, S9). Surprisingly however, further removal of miR-58 members caused a greater increase in RNA than protein abundance, resulting in an apparent reduction in target translation efficiency in the quadruple mutant. This effect originates from the targets expressed in the germline (Supplemental Fig. S11), indicating tissue-specific differences in miRNA target regulation, reminiscent of a recent zebrafish study (Subtelny et al. 2014). One possible explanation for this counterintuitive observation is that in these mutants, a fraction of miR-58 family targets, albeit rescued from mRNA degradation, are not licensed for translation, for example, because they remain in P granules or P bodies. Alternatively, the miR-58 family might compete with a hypothetical RNA-binding protein that also recognizes the miR-58 family seed sequence and that might act mainly at the level of translational repression. Altogether, the comparative analysis of target protein and RNA abundance changes in the miR-58 family mutants in the germline and in the soma indicate that the miR-58 family members down-regulate their targets mainly by inducing mRNA degradation, consistent with previous reports (Hendrickson et al. 2009; Guo et al. 2010; Stadler et al. 2012). As our analysis was restricted to the whole animals in the fourth larval stage of development, it might miss revealing some tissue- or stage-specific mechanistic aspects of miRNA repression, for example in the embryos (Wu et al. 2010). Furthermore, since we measured steady-state protein and mRNA levels, we cannot exclude the possibility that the effects on protein stability and mRNA metabolism could have had an impact on our measurements.
Conclusions
In summary, in this study we undertook a systematic approach to explore target regulation by miR-58 family members in C. elegans. We demonstrate that the miRNAs exert a cumulative effect on largely overlapping target sets, and show that although some effects on translation can be observed, the predominant mechanism of miRNA-induced repression is, at least at the aggregate level, through target mRNA degradation. It will be important to expand such studies to mammalian miRNA families to identify the overlapping target sets of miRNA family members and the exact levels of target alterations that can lead to disease. Several miRNA families are oncogenic, their members being simultaneously up-regulated in cancers (He et al. 2005; Mi et al. 2010; Liu et al. 2013). Many of these families likely cooperate to regulate their targets. For this reason, it will be essential to further advance approaches sequestering multiple family members. Recently developed locked nucleic acid (LNA)–modified antimiRs have been successfully applied to target miRNA families in mammalian cell lines, mice, and primates (Obad et al. 2011; Rottiers et al. 2013) and opened space for clinical applications of miRNA family inhibition. Given the omnipresence of miRNA families in the animal kingdom and their widespread impact on development, homeostasis, and disease, we envisage that further research on target regulation mediated by miRNA family members will pave the way to understanding of a large number of biological processes.
Methods
C. elegans strains and culture conditions
C. elegans strains were maintained as described previously (Brenner 1974). All strains were raised at 20°C or 25°C. The Bristol N2 strain was utilized as the wild type. The following miR-58 family mutations were used in this study: LGIII, mir-80(nDf53); LGIV, mir-81-82(nDf54); and LGX, mir-58.1(n4640). The three mutants and all their combinations, which were previously generated and described (Alvarez-Saavedra and Horvitz 2010), were obtained from the Caenorhabditis Genetics Center and the laboratory of Prof. Robert Horvitz at the Massachusetts Institute of Technology. The transgenes (Alvarez-Saavedra and Horvitz 2010) nEx1457 [miR-80], nEx1581 [miR-58.1], nEx1582 [T07D1.2::miR-82], and nEx1610 [ΔmiR-80] were used to test the rescue of defective IR-induced apoptosis of quadruple mutants.
Stable isotope labeling by amino acids in nematodes
Heavy- and light-labeled samples of the genotypes—wild type, mir-58.1, mir-80; mir-58.1, and mir-80; mir-58.1; mir-81-82—were prepared by feeding with heavy- and light-labeled bacteria, respectively. Worms were harvested at the late L4 stage for subsequent protein and RNA isolation. Three biological replicates were grown, and the experiment was performed in two variants: one without fractionation (in three biological replicates) and the other with HILIC fractionation (one replicate). Proteins from all the samples were extracted with 50 mM Tris/HCl (pH 8.3), 5 mM EDTA, 8 M urea buffer, and glass beads as described previously (Schrimpf et al. 2009). Peptides obtained from SILAC samples were fractionated with a Agilent 1100 HPLC flow system using a 4-mm HILIC column YMC-pack polyamine II (YMC Europe GmbH). Samples were analyzed by LC-MS/MS using a Thermo easy nLC 1000 HPLC system coupled to a Thermo Orbitrap elite hybrid mass spectrometer equipped with a Nanoflex electrospray source (Thermo Fisher Scientific). Further details on SILAC sample preparation, protein isolation, HILIC fractionation, and mass spectrometric measurements can be found in the Supplemental Methods.
Selected reaction monitoring
Wild type, mir-58.1, mir-80, mir-81-82, mir-80; mir-58.1, mir-58.1; mir-81-82, mir-80; mir-81-82, and mir-80; mir-58.1; mir-81-82 were grown in three biological replicates for targeted proteomics measurement. Due to the developmental delay of the mir-58.1; mir-81-82 triple mutant, mir-80; mir-58.1 double mutant, and mir-80; mir-58.1; mir-81-82 quadruple mutant, worms were collected 3, 6, and 14 h after the wild type (and other miR-58 family mutants that showed no developmental delay at 25°C: mir-80, mir-81-82, mir-80; mir-81-82, and mir-58.1), respectively. Previously developed SRM assays (Jovanovic et al. 2010, 2012) for 24 proteins predicted to be miR-58 targets by TargetScan and a control group of 22 random proteins were used on all combinations of miR-58 family mutants. Measurements were performed on a Thermo Quantum ultra triple quadrupole mass spectrometer. For a detailed description of the measurements and statistical analysis, see the Supplemental Methods.
Transcriptome sequencing
Transcriptome sequencing of RNA isolated from the wild type, mir-58.1 single, mir-80; mir-58.1 double, and mir-80; mir-58.1; mir-81-82 quadruple mutants in three biological replicates was performed in GATC Biotech. The differential expression analysis was performed using DESeq (Anders and Huber 2010), comparing the number of reads in three replicates of one condition with those in the replicates of another condition.
Quantitative real-time PCR on miR-58 family targets and egl-1 induction
Twenty nanograms of RNA was used for cDNA synthesis with the SuperScript III (Invitogen) as instructed by the manufacturer. qRT-PCR for miR-58 targets was performed in two technical and three biological replicates and egl-1 qRT-PCR in three technical and two biological replicates. The mean measured levels for transcripts of interest were normalized to the mean of a set of internal controls: housekeeping genes pgk-1, mpk-1, and tbp-1 and two additional reliable reference genes, cdc-42 and Y45F10D.4 (Hoogewijs et al. 2008). Primers used for qRT-PCR are listed in Supplemental Table 5, and additional details on sample preparation and processing are given in the Supplemental Methods.
Small RNA sequencing
One microgram of total RNA isolated with TRIzol according to the manufacturer's protocol was processed for small RNA sequencing (for extensive description of library preparation, see the Supplemental Methods). Four calibrator oligonucleotides (Cal 01-04, 5 fMol each) were added as a reference as described previously (Hafner et al. 2012a, b). RNA was dephosphorylated using FastAP (Fermentas) and radiolabeled according to the described method (Hafner et al. 2012a) with the 10-fold reduction of γ-32P-ATP (final concentration, 0.05 μCi/μL).
miRNA qRT-PCR
miR-58 family member quantification in late L4 staged animals was carried out using an adaptation of the stem–loop qRT-PCR protocol (Chen et al. 2005). Protocol details can be found in the Supplemental Methods. Primers are listed in Supplemental Table 5.
Data access
The RNA-sequencing and small RNA sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE60421. SILAC data are available via ProteomeXchange with identifier PXD001998 (http://www.ebi.ac.uk/pride).
Supplementary Material
Acknowledgments
Many experiments reported in this study were performed in the Functional Genomics Center Zurich (FGCZ). We thank B. Roschitzki, A. Wahlander, and C. Fortes from the FGCZ for the technical support with TSQ Quantum Ultra, Agilent HPLC 1100 and sample preparation; Alexander Leitner for the technical support with Orbitrap Elite; Martin Moser for the technical support with qRT-PCR; Michael Daube for help in the generation of transgenic strains; Kapil Singh for the statistical support; Robert Horvitz for the MT14128, MT18091, MT18755, MT18756, and MT18999 strains; and members of the Hengartner laboratory for the critical discussions on the project. Some miR-58 family mutant strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work was funded in part by Swiss National Science Foundation Sinergia grant CRSII3_141942, University of Zurich Research Priority Program in Systems Biology and the ETH Zurich. S.G. was supported by a grant of the Swiss National Science Foundation (Marie-Heim Voegtlin, PMPDP3_122836). S.N. was supported by the Seventh Framework Program of the European Union (contract no. 262067-PRIME-XS).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.183160.114.
References
- Alvarez-Saavedra E, Horvitz HR. 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol 20: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature 455: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazie SM, Babb C, Wilky H, Rawls A, Park JG, Mangone M. 2015. Comparative RNA-Seq analysis reveals pervasive tissue-specific alternative polyadenylation in Caenorhabditis elegans intestine and muscles. BMC Biol 13: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. 2007. microRNA functions. Annu Rev Cell Dev Biol 23: 175–205. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. 2006. MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. 2005. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Hannon GJ, Darnell RB. 2012. An alternative mode of microRNA target recognition. Nat Struct Mol Biol 19: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepcion CP, Bonetti C, Ventura A. 2012. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J 18: 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B, Horvitz H. 1998. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2–like protein CED-9. Cell 93: 519–529. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard R, Stergiou L, Hofmann ER, Hofmann J, Haenni S, Teo Y, Furger A, Hengartner MO. 2013. Ribosome synthesis and MAPK activity modulate ionizing radiation-induced germ cell apoptosis in Caenorhabditis elegans. PLoS Genet 9: e1003943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. 2010. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79: 351–379. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ. 2008. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle 7: 3112–3117. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N. 2014. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol Cell 54: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Lianoglou S, Tuschl T, Betel D. 2012a. Genome-wide identification of miRNA targets by PAR-CLIP. Methods 58: 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Renwick N, Farazi TA, Mihailović A, Pena JT, Tuschl T. 2012b. Barcoded cDNA library preparation for small RNA profiling by next-generation sequencing. Methods 58: 164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser J, Syed AP, Selevsek N, van Nimwegen E, Jaskiewicz L, Aebersold R, Zavolan M. 2013. Timescales and bottlenecks in miRNA-dependent gene regulation. Mol Syst Biol 9: 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. 2005. A microRNA polycistron as a potential human oncogene. Nature 435: 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. 2013. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. 2009. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol 7: e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. 2008. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu C, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65: 7065–7070. [DOI] [PubMed] [Google Scholar]
- Isik M, Korswagen HC, Berezikov E. 2010. Expression patterns of intronic microRNAs in Caenorhabditis elegans. Silence 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′ UTRs. Nature 469: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M, Reiter L, Picotti P, Lange V, Bogan E, Hurschler BA, Blenkiron C, Lehrbach NJ, Ding XC, Weiss M, et al. 2010. A quantitative targeted proteomics approach to validate predicted microRNA targets in C. elegans. Nat Methods 7: 837–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M, Reiter L, Clark A, Weiss M, Picotti P, Rehrauer H, Frei A, Neukomm LJ, Kaufman E, Wollscheid B, et al. 2012. RIP-chip-SRM: A new combinatorial large-scale approach identifies a set of translationally regulated bantam/miR-58 targets in C. elegans. Genome Res 22: 1360–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, de Lencastre A, Pincus Z, Slack FJ. 2009. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol 10: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorshid M, Hausser J, Zavolan M, van Nimwegen E. 2013. A biophysical miRNA–mRNA interaction model infers canonical and noncanonical targets. Nat Methods 10: 253–255. [DOI] [PubMed] [Google Scholar]
- Kim Y, Wee G, Park J, Kim J, Baek D, Kim J, Kim VN. 2013. TALEN-based knockout library for human microRNAs. Nat Struct Mol Biol 20: 1458–1464. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlow BA, Zhang L, Han M. 2012. Systematic analysis of tissue-restricted miRISCs reveals a broad role for microRNAs in suppressing basal activity of the C. elegans pathogen response. Mol Cell 46: 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. 2009. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′ UTR microRNA recognition elements. Mol Cell 35: 610–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Hook B, Pan G, Kershner AM, Merritt C, Seydoux G, Thomson JA, Wickens M, Kimble J. 2007. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet 3: e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Liu N, Okamura K, Tyler DM, Phillips MD, Chung W, Lai EC. 2008. The evolution and functional diversification of animal microRNA genes. Cell Res 18: 985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Nie J, Chen L, Dong G, Du X, Wu X, Tang Y, Han W. 2013. The oncogenic role of microRNA-130a/301a/454 in human colorectal cancer via targeting Smad4 expression. PLoS One 8: e55532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. 2012. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell 48: 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. 2007. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci 104: 9667–9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciotta S, Meregalli M, Torrente Y. 2013. The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci 7: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, Walhout AJM. 2008. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res 18: 2005–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Olson EN. 2012. MicroRNAs in stress signaling and human disease. Cell 148: 1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Li Z, Chen P, He C, Cao D, Elkahloun A, Lu J, Pelloso LA, Wunderlich M, Huang H, et al. 2010. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc Natl Acad Sci 107: 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. 2007. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet 3: e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nana-Sinkam SP, Croce CM. 2011. Non-coding RNAs in cancer initiation and progression and as novel biomarkers. Mol Oncol 5: 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al. 2011. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet 43: 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano DJ, Kingston ER, Kim DH. 2015. Tissue expression pattern of PMK-2 p38 MAPK is established by the miR-58 family in C. elegans. PLoS Genet 11: e1004997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Gaur AB, Lengyel E, Peter ME. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22: 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Lee JH, Ha M, Nam J, Kim VN. 2009. miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nat Struct Mol Biol 16: 23–29. [DOI] [PubMed] [Google Scholar]
- Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu C, Rassenti L, et al. 2006. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res 66: 11590–11593. [DOI] [PubMed] [Google Scholar]
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TLJ, Visakorpi T. 2007. MicroRNA expression profiling in prostate cancer. Cancer Res 67: 6130–6135. [DOI] [PubMed] [Google Scholar]
- Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, Black JC, Sinha S, Goody RJ, Lawrence MS, deLemos AS, et al. 2013. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med 5: 212ra162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski R, Dickinson R, Stewart G, Craig A, Schimpl M, Keyse SM, Gartner A. 2011. Regulation of Caenorhabditis elegans p53/CEP-1-dependent germ cell apoptosis by Ras/MAPK signaling. PLoS Genet 7: e1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrimpf SP, Weiss M, Reiter L, Ahrens CH, Jovanovic M, Malmström J, Brunner E, Mohanty S, Lercher MJ, Hunziker PE, et al. 2009. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol 7: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Hofmann K, Boulton S, Gartner A. 2001. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr Biol 11: 1722–1727. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Schertel C, Wittenburg N, Tuck S, Mitani S, Gartner A, Conradt B, Shaham S. 2005. C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ 12: 153–161. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63. [DOI] [PubMed] [Google Scholar]
- Shaw WR, Armisen J, Lehrbach NJ, Miska EA. 2010. The conserved miR-51 microRNA family is redundantly required for embryonic development and pharynx attachment in Caenorhabditis elegans. Genetics 185: 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Nam J, Farh KK, Chiang HR, Shkumatava A, Bartel DP. 2010. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell 38: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M, Artiles K, Pak J, Fire A. 2012. Contributions of mRNA abundance, ribosome loading, and post- or peri-translational effects to temporal repression of C. elegans heterochronic miRNA targets. Genome Res 22: 2418–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Choi E, Lin S, Reinert K, Slack FJ. 2004a. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′ UTR. Genes Dev 18: 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Reinert K, Slack FJ. 2004b. Architecture of a validated microRNA::target interaction. Chem Biol 11: 1619–1623. [DOI] [PubMed] [Google Scholar]
- Wang Q, Wei L, Guan X, Wu Y, Zou Q, Ji Z. 2014. Briefing in family characteristics of microRNAs and their applications in cancer research. Biochim Biophys Acta 1844: 191–197. [DOI] [PubMed] [Google Scholar]
- Wu E, Thivierge C, Flamand M, Mathonnet G, Vashisht AA, Wohlschlegel J, Fabian MR, Sonenberg N, Duchaine TF. 2010. Pervasive and cooperative deadenylation of 3′ UTRs by embryonic microRNA families. Mol Cell 40: 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. 2006. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189–198. [DOI] [PubMed] [Google Scholar]
- Yang Y, Chaerkady R, Kandasamy K, Huang T, Selvan LD, Dwivedi SB, Kent OA, Mendell JT, Pandey A. 2010. Identifying targets of miR-143 using a SILAC-based proteomic approach. Mol Biosyst 6: 1873–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.