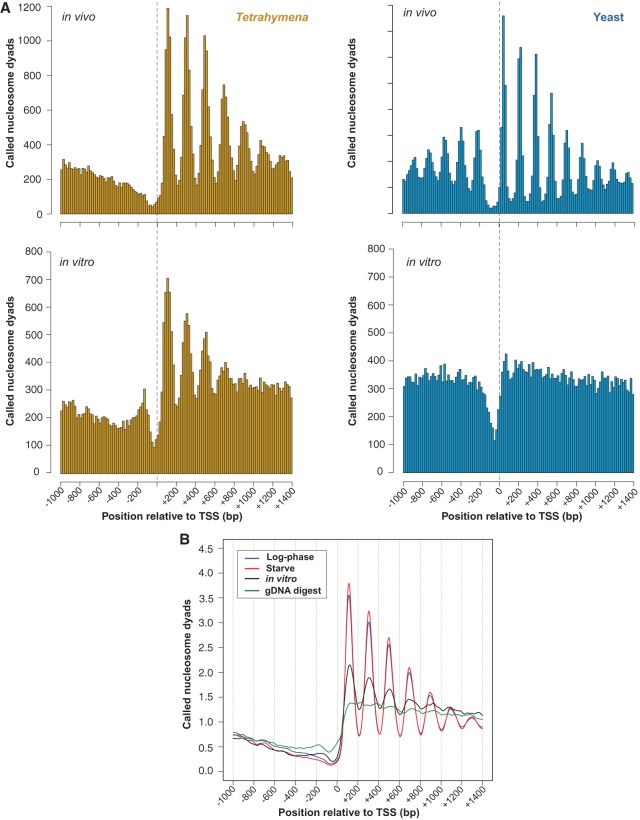
Figure 1.
In vivo-like nucleosome organization without trans-acting factors. (A) Histograms of nucleosome positions relative to the TSS were computed from yeast and Tetrahymena MNase-seq data using the same bioinformatic pipeline. A phased distribution of nucleosome positions downstream from the TSS is observed in chromatin from log-phase Tetrahymena and yeast grown in rich media. Surprisingly, an in vivo-like pattern of nucleosome positioning is observed in vitro for Tetrahymena but not yeast. (B) Averaged nucleosome dyad counts around the TSS reveal an in vivo-like distribution of called nucleosomes within in vitro data. MNase-digested naked DNA does not resemble in vivo data (green curve), thus ruling out potential sequence biases associated with MNase preferences.