Figure 1.
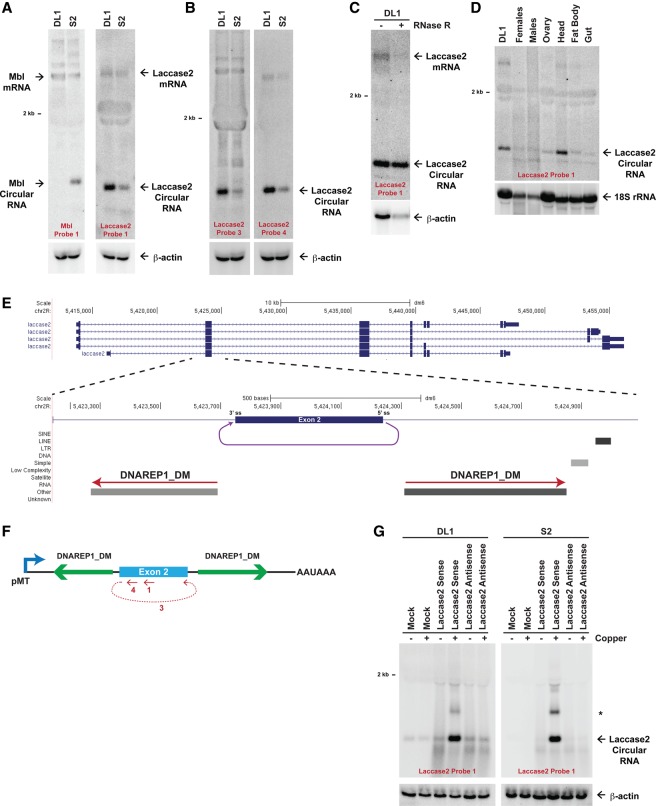
The D. melanogaster laccase2 gene generates a circular RNA. (A) Twenty micrograms of total RNA from DL1 and S2 cells was subjected to Northern blot analysis and probed for Mbl and Laccase2 expression. β-Actin was used as a loading control. (B) The Laccase2 circular RNA was detected with multiple oligonucleotide probes, including one complementary to the backspliced junction (probe 3). (C) The Laccase2 circular RNA is resistant to RNase R digestion. (D) Eleven micrograms of total RNA from adult D. melanogaster tissues was probed for Laccase2 expression. 18S ribosomal RNA was used as a loading control. (E) Exon/intron structure of the D. melanogaster laccase2 locus, highlighting a 1945-nt region that includes exon 2. A circular RNA is formed when the 5′ splice site at the end of exon 2 is joined to the 3′ splice site at the beginning of exon 2 (purple). Repetitive elements in the designated orientations are shown. (F) The 1945-nt region of the Laccase2 pre-mRNA was cloned downstream from the metallothionein promoter to generate the Laccase2 sense plasmid. The regions targeted by Northern oligonucleotide probes are denoted in red. (G) Plasmids containing the laccase2 region in the sense or antisense orientation were transfected into DL1 (left) or S2 (right) cells, and Northern blots were performed. Endogenous Laccase2 circular RNA expression was observed in the mock-treated samples. (*) Concatenated and/or intertwined circular RNA. β-Actin was used as a loading control.