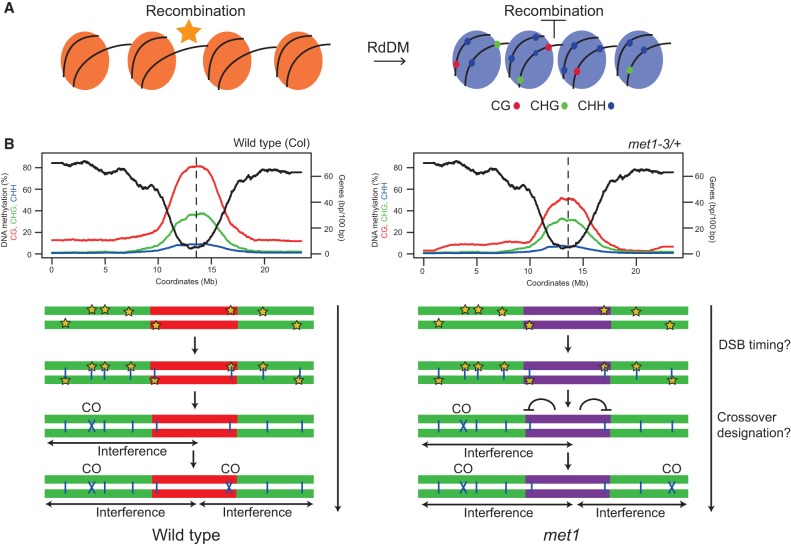
Figure 7.
Control of meiotic recombination by DNA methylation in Arabidopsis. (A) Epigenetic changes associated with RdDM at crossover hot spots. Initially, the hot spot has low DNA methylation, low H3K9me2 (orange nucleosomes), and low nucleosome occupancy and is permissive to recombination. Following establishment of RdDM, dense DNA methylation in CG (red), CHG (green), and CHH (blue) sequence contexts is acquired in addition to higher nucleosome occupancy and H3K9me2 (blue nucleosomes), together causing recombination suppression. (B) The top plots show gene density (base pairs per 100 bp, black) and percentage of DNA methylation (CG in red, CHG in green, and CHH in blue) along chromosome 3 in wild type (Col) and met1-3/+. (Bottom) Chromosome representations with euchromatin colored green and heterochromatin colored red (wild type) or purple (met1). Meiotic DSBs (stars) form in a similar pattern along the chromosome in wild type and met1. DSBs are resected to form ssDNA that invades the homologous chromosome (blue vertical lines). A subset of interhomolog invasion events become crossover-designated and impose interference on adjacent unrepaired DSBs. We propose that loss of CG methylation in the centromeric regions in met1 either (1) alters the relative timing of DSB formation between euchromatin and heterochromatin or (2) reduces the chance of interhomolog invasion events becoming crossover-designated in proximity to the centromere. Interference operates normally in met1, and, as a consequence, the euchromatic arms receive additional crossovers compared with wild type.