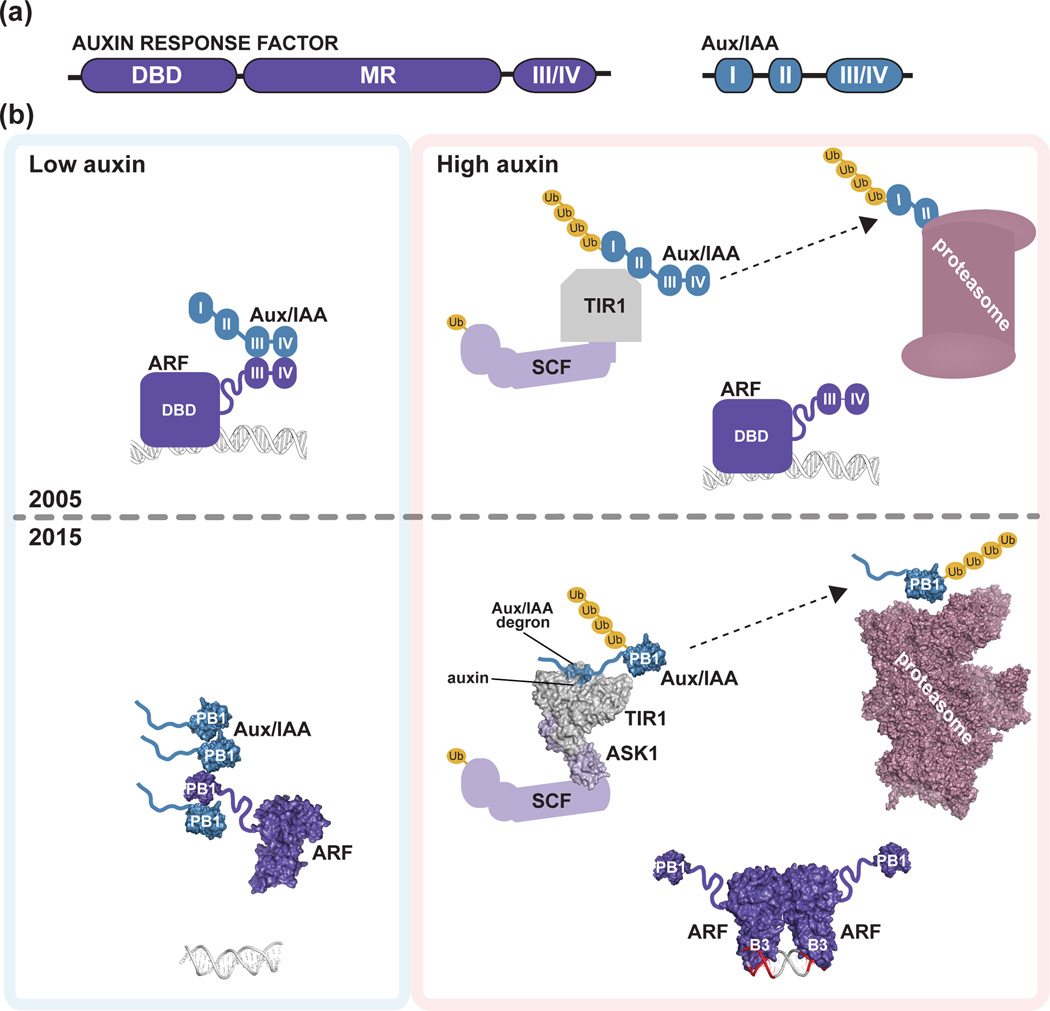
Figure 1. Initial and refined auxin response models.
(a) Schematic of homology motifs in Auxin Response Factor (ARF) and Aux/IAA proteins. The ARF DNA binding domain (DBD), middle region (MR), and III/IV interaction motif are indicated. The Aux/IAA sequence motifs I (TOPLESS interaction), II (degron), and III/IV (interaction motif) are shown. (b) The top panel depicts the auxin response model from the year 2005. Under low local auxin concentrations, Aux/IAA proteins interact with ARF proteins, thereby repressing their action. In the presence of auxin , TIR1/AFB perceives auxin, allowing for SCFTIR1/AFB interaction with Aux/IAA proteins. Subsequently, Aux/IAA repressors are degraded via the 26S proteasome to allow for ARF-mediated gene expression changes. The bottom panel integrates structural biology contributions to our understanding of auxin signaling, including the ‘molecular glue’ interaction of auxin with TIR1 that allows for interaction of SCFTIR1/AFB with Aux/IAA repressors (PDB: 2P1Q; [22]); the ARF5 DBD structure (PDB: 4LDU; [34]) providing the ‘molecular calipers’ model of AuxRE recognition (PDB: 4LDX; [34]), and the possibility of PB1 domain multimerization in ARF (PDB: 4NJ6; [39]) and Aux/IAA (PDB: 2MUK; [42]) proteins to regulate auxin signaling. The cryo-EM proteasome structure is also shown for reference ([49]; PDB: 4CR2).