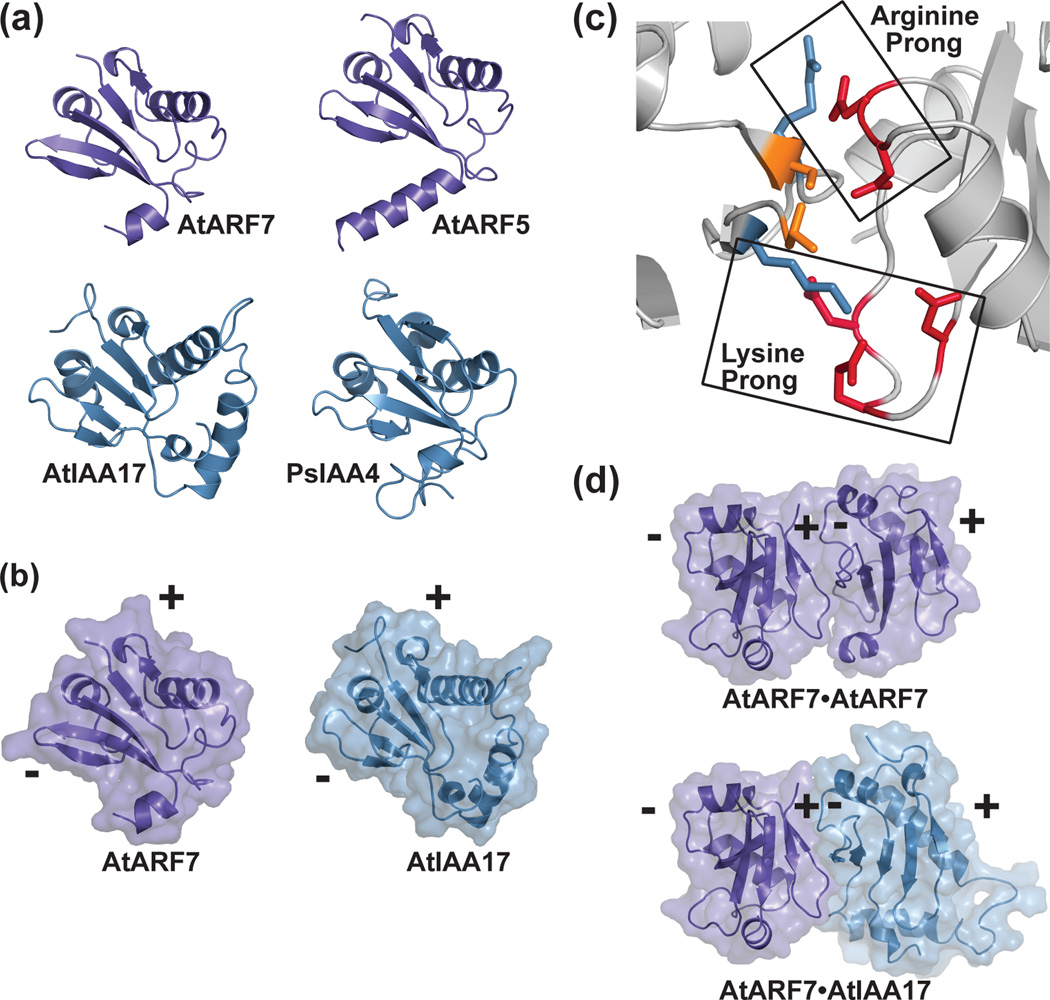
Figure 4. Crystal structures of the ARF and Aux/IAA PB1 domains and the versatility of protein-protein interaction.
(a) The X-ray crystal structures of the PB1 domains of AtARF7 (PDB: 4NJ6; [39]) and AtARF5 (PDB: 4CHK; [40]) and the NMR solution structures of AtIAA17 (PDB: 2MUK; [42]) and PsIAA4 (PDB: 2MIM) reveal a common fold. (b) The PB1 domain scaffold presents conserved residues on opposing positive (+) and negative (-) interaction surfaces. A mixed surface/ribbon view of the PB1 domains of AtARF7 [39] and AtIAA17 [42] are shown. (c) PB1-mediated interaction of ARF and/or Aux/IAA proteins use a two-pronged binding motif. Biophysical studies of the ARF7 PB1 domain (PDB: 4NJ6; [39]) revealed how conserved lysine and arginine residues on the positive-face interact with two structurally distinct groups of acidic residues. (d) PB1 domains of ARF and Aux/IAA provide a versatile scaffold for protein interaction. Mixed surface/ribbon diagram showing the crystallographically-observed head-to-tail orientation of two AtARF7 PB1 domains (PDB: 4NJ6; [39]). Modeling of a hypothetical AtARF7•AtIAA17 complex, based on the AtARF7•AtARF7 structure, predicts the overall scaffold allowing for mixed-protein interaction.