Abstract
ARGONAUTES (AGOs) are the effector proteins functioning in eukaryotic RNA silencing pathways. AGOs associate with small RNAs and are programmed to target complementary RNA or DNA. Plant viruses induce a potent and specific antiviral RNA silencing host response in which AGOs play a central role. Antiviral AGOs associate with virus-derived small RNAs to repress complementary viral RNAs or DNAs, or with endogenous small RNAs to regulate host gene expression and promote antiviral defense. Here, we review recent progress towards understanding the roles of plant AGOs in antiviral defense. We also discuss the strategies that viruses have evolved to modulate, attenuate or suppress AGO antiviral functions.
Introduction
ARGONAUTES (AGOs) are the effector proteins functioning in eukaryotic RNA silencing pathways [1]. Plant AGOs associate with small RNA based on the identity of the 5′ terminal nucleotide of the small RNA and/or other sequence and structural properties of the small RNA duplex [2]. Small RNA-programmed AGOs target and silence complementary RNA or DNA through posttranscriptional gene silencing (PTGS) or transcriptional gene silencing (TGS), respectively [3].
RNA silencing functions as a primary antiviral immune system in plants [4]. Antiviral RNA silencing is triggered by highly-structured or double-stranded (ds) viral RNAs that are recognized and processed by host Dicer-Like (DCL) proteins into primary 21 to 24 nt virus-derived small interfering RNAs (vsiRNAs). Endogenous RNA-dependent RNA polymerases (RDRs) use single-stranded (ss) viral RNAs to synthesize dsRNA serving as substrate for DCLs to produce secondary vsiRNAs and amplify the antiviral response. Antiviral AGOs associate with vsiRNAs and i) target complementary viral RNA for degradation through endonucleolytic cleavage (slicing) and/or translational arrest, ii) transcriptionally repress complementary viral DNA through hypermethylation, or iii) regulate host gene expression to promote defense [4]. As a counter-defense strategy, most if not all plant viruses have evolved specialized proteins known as viral suppressors of RNA silencing (VSRs) [5•]. VSRs can interfere with virtually any step of the antiviral RNA silencing pathway, and attenuate or completely suppress the defense response.
In this article, we review recent progress towards understanding which AGOs have antiviral roles in plants, and how they operate through association with vsiRNAs or through other mechanisms. We also discuss how viruses affect AGOs to regulate host gene expression and promote viral infection, and how they suppress AGO antiviral functions in this molecular arms race between plants and viruses.
Multiple AGOs function in plant antiviral silencing
The Arabidopsis thaliana (Arabidopsis) genome encodes 10 AGO genes [6]. Early genetic analyses describing Arabidopsis ago1 hypersusceptibility to Cucumber mosaic virus (CMV) revealed the first antiviral role for a plant AGO [7]. Later, the suspicion that functional VSRs could mask the effects of antiviral silencing functions prompted the use of suppressor-defective viruses to analyze ago activity, and AGO1 was identified as the major antiviral AGO against suppressor-defective Turnip crinkle virus (TCV) in Arabidopsis [8]. Similar analyses of Arabidopsis single or combinational ago mutants with different wild-type and/or VSR-defective viruses confirmed AGO1 [9–11•] and AGO2 [9,11•–16] as two major antiviral AGOs against RNA viruses in Arabidopsis. It was initially reported that AGO2 could function in cooperation, and non-redundantly, with AGO1 in defense against suppressor-defective CMV [9], acting as a second defense layer against viruses that suppress AGO1 function (e.g. CMV or TCV) [12]. It was also hypothesized that the decrease of AGO1 levels upon virus infection induces the accumulation of AGO2 as a result of reduced AGO2 targeting by AGO1/miR403 complexes [12]. More recent observations have confirmed that AGO2 is indeed the main antiviral AGO against other viruses that are not known to target AGO1 such as Tobacco rattle virus (TRV) [16] and Turnip mosaic virus (TuMV) [11]. Other AGOs acting in PTGS, such as AGO5, AGO7 and AGO10, have been genetically and/or biochemically implicated in antiviral defense but with minor roles [8,11•,17]. Interestingly, a modular anti-TuMV activity of several Arabidopsis AGOs in various organs [11•] suggested that antiviral AGOs may work coordinately and cooperatively in a spatiotemporal manner. Finally, genetic and biochemical evidences support that AGO4, functioning in TGS, plays an antiviral role in Arabidopsis against several RNA [16,18,19] and DNA viruses [20,21].
The contribution of AGOs in antiviral silencing in species other than Arabidopsis was not examined until recently. In Nicotiana benthamiana, AGO2 protects against suppressor-defective Tomato bushy stunt virus (TBSV) [22], while AGO1 is required for temperature-dependent symptom recovery against Tomato ringspot virus (ToRSV) [23]. In rice, genetic and/or biochemical evidences suggest that AGO1 and AGO18 are the main antiviral AGOs against Rice stripe tenuivirus (RSV) and Rice dwarf phytoreovirus (RDV) [24••]. Table 1 lists the specific plant AGOs known to have antiviral activity.
Table 1.
Plant AGOs with antiviral activity.
| AGO | Virusa | Host(s) | Reference(s) |
|---|---|---|---|
| AGO1 | BMV | Arabidopsis thaliana | [10] |
| CMV | Arabidopsis thaliana | [7] | |
| CMV | Arabidopsis thaliana | [9] | |
| RDV | Oryza sativa | [24] | |
| RSV | Oryza sativa | [24] | |
| ToRSV | Nicotiana benthamiana | [23] | |
| TCV | Arabidopsis thaliana | [8] | |
| TuMV | Arabidopsis thaliana | [11] | |
| AGO2 | CMV | Arabidopsis thaliana | [9, 12, 17] |
| PVX | Arabidopsis thaliana | [13] | |
| TRV | Arabidopsis thaliana | [16] | |
| TBSV | Nicotiana benthamiana | [22] | |
| TCV | Arabidopsis thaliana | [12, 15] | |
| TuMV | Arabidopsis thaliana | [11, 14] | |
| AGO4 | BCTV | Arabidopsis thaliana | [20] |
| CaMV | Arabidopsis thaliana | [21] | |
| CMV | Arabidopsis thaliana | [18] | |
| PVX | Nicotiana benthamiana | [19] | |
| TRV | Arabidopsis thaliana | [16] | |
| AGO5 | CMV | Arabidopsis thaliana | [17] |
| TuMV | Arabidopsis thaliana | [11] | |
| AGO7 | TCV | Arabidopsis thaliana | [8] |
| TuMV | Arabidopsis thaliana | [11] | |
| AGO10 | TuMV | Arabidopsis thaliana | [11] |
| AGO18 | RDV | Oryza sativa | [24] |
| RSV | Oryza sativa | [24] |
BCTV, Beet curly top virus; BMV, Brome mosaic virus; CaMV, Cauliflower mosaic virus; CMV, Cucumber mosaic virus; PVX, Potato virus X; RDV, Rice dwarf phytoreovirus; RSV, Rice stripe tenuivirus; TRV, Tobacco rattle virus; TBSV, Tomato bushy stunt virus; ToRSV, Tomato ringspot virus; TCV, Turnip crinkle virus; TuMV, Turnip mosaic virus.
Antiviral functions of AGOs programmed with virus-derived small RNAs
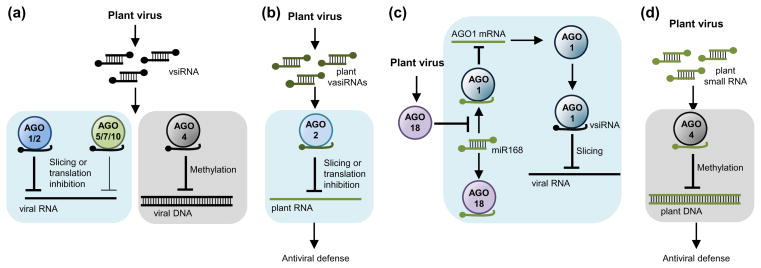
Antiviral AGOs associating with vsiRNAs are typically programmed to target and repress cognate viral RNA or DNA (Figure 1a). AGO association with vsiRNAs upon virus infection has been reported for Arabidopsis AGO1 [9,11•,15], AGO2 [9,11•,12,17], AGO5 [17] and AGO10 [11•], and for rice AGO1 and AGO18 [24••].
Figure 1.
ARGONAUTE functions in plant antiviral RNA silencing. (a) AGOs programmed with vsiRNAs repress viral RNA through slicing or translation inhibition in PTGS, or through hypermethylation of viral DNA in TGS. AGO1 and AGO2 are the main antiviral AGOs against RNA viruses, with AGO5, AGO7 and AGO10 having minor roles in some cases. AGO4 is the main antiviral AGO against DNA viruses. (b–d) AGOs programmed with plant small RNAs promote antiviral defense. (b) In Arabidopsis, virus infection triggers the production of endogenous vasiRNAs that work mainly through AGO2 to repress transcripts from genes mostly related to response to biotic and abiotic stimuli and ribosomal RNA. (c) In rice, plant virus infection induces the accumulation of AGO18 which sequesters miR168 away from AGO1, and consequently induces AGO1 accumulation at elevated levels necessary for antiviral defense. (d) In infected Arabidopsis, AGO4 can bind to plant small RNAs to regulate host transcriptional response to infection and promote antiviral defense. Light blue and grey boxes include steps in PTGS or TGS, respectively.
ARGONAUTES loaded with vsiRNA complexes can form antiviral RNA-induced silencing complexes (RISCs) that target complementary viral RNAs for degradation through slicing or translational arrest [4]. Early evidence supporting the existence of endonucleolytic activity of antiviral RISCs was gathered from in vitro experiments involving different combinations of cell-free systems and viruses [4]. However, the involvement of an AGO protein as part of the active RISC was not demonstrated until later experiments using cytoplasmic extracts of evacuolated tobacco protoplasts to test the AGO-mediated cleavage of synthetic [25] or replicating target RNAs [26•]. In evacuolated protoplasts transfected with replicating TBSV, RISCs containing AGO1 and AGO2 strongly inhibited TBSV replication and targeted neighboring sequences, suggesting that some regions of the viral RNAs could be more accessible and therefore collectively targeted by multiple AGOs [26•]. In vivo evidence of AGO-mediated cleavage of viral RNAs was initially limited to the genetic involvement of AGO1 and AGO7 in the clearance of highly and less structured, respectively, TCV-based sensor RNAs in Arabidopsis [8], and to the targeting of the 3′ untranslated region of CMV by a satellite-derived siRNA associating with AGO1 in N. benthamiana [27]. More recently, AGO2 slicing activity was shown to be critical for antiviral resistance in suppressor-defective TuMV infected Arabidopsis [14]. Because AGO2 binds suppressor-defective TuMV-derived vsiRNAs [11•], it is possible that AGO2 slices viral RNAs in vivo. Finally, AGO/vsiRNA complexes could target viral RNA through slicing-independent mechanisms as proposed to explain the AGO1-dependent translational arrest of ToRSV RNA2 to promote temperature-dependent symptom recovery in N. benthamiana [23].
AGO/vsiRNA complexes can also form antiviral RNA-induced transcriptional silencing complexes (RITS) that target viral DNA. AGO4-dependent hypermethylation of viral DNA in Arabidopsis was first observed during host recovery from Beet curly top virus (BCTV) infection [20], and more recently it was shown to require DOUBLE-STRANDED RNA-BINDING PROTEIN 3 during Cabbage leaf curl virus (CaLCuV) and BCTV infections [21].
Antiviral functions of AGOs programmed with host small RNAs
AGOs can also associate with endogenous small RNAs to regulate host gene expression and promote antiviral defense (Figure 1b–d). In Arabidopsis, AGO4 was proposed to participate in anti-CMV defense by regulating host transcriptional response during infection through its association with endogenous small RNAs [18]. More recently, CMV infection was shown to induce the synthesis of an abundant class of endogenous 21 nt siRNAs that mapped to the exon regions of more than 1000 Arabidopsis genes (most of them known to respond to biotic and abiotic stimuli) and ribosomal RNA (rRNA) [28••]. This new class of endogenous siRNAs, named virus-activated siRNAs (vasiRNAs), are DCL4/RDR1-dependent, function mainly through AGO2, and enhance antiviral resistance. As vasiRNA accumulation was also triggered by TuMV, a virus belonging to a different supergroup, it was proposed that the production of vasiRNAs may provide a broad-spectrum antiviral activity complementary to the virus-specific resistance directed by vsiRNAs [28••]. However, the role of rRNA-derived vasiRNAs is still unclear, as these are not specifically loaded into AGO1 or AGO2 and do not seem to direct RNA silencing in infected plants. It is not obvious either how to explain the absence of silencing of host genes by AGO1/CMV-vasiRNA complexes, or the suppression of vasiRNA biogenesis by CMV 2b and not by TuMV HC-Pro despite that ability of both to sequester siRNAs. In rice, infection with RSV and RDV triggers AGO18 accumulation [24]. As observed in RSV and RDV infections, AGO18 interferes with AGO1 homeostatis by sequestering miR168 from AGO1, and consequently inducing AGO1 accumulation at elevated levels required for antiviral defense [24].
Virus-derived small RNAs hijack AGOs to target host transcripts
Plant viruses can use host RNA silencing factors to facilitate their infection (Figure 2a). For example vsiRNAs can hijack AGOs to target sufficiently complementary host transcripts, as reported in two parallel studies that linked the targeting of CHLI (gene involved in chlorophyll synthesis) transcripts by siRNAs derived from the Y satellite of CMV with characteristic yellow symptoms in infected tobacco plants [29,30]. Similarly, the albino phenotype characteristic of peach leaves infected with Peach latent mosaic viroid (PLMVd) is due to the specific down-regulation of host CHLOROPLASTIC HEAT-SHOCK PROTEIN 90 transcripts by two PLMVd-derived siRNAs [31]. More recently, the parallel analysis of several vsiRNA and viral RNA degradome high-throughput sequencing datasets from virus-infected grape plants identified a pool of vsiRNAs predicted to target host transcripts, some of which encode proteins involved in ribosome biogenesis and biotic stress [32]. However, only a small subset of these transcripts were validated as putative targets by 5′ Rapid Amplification of cDNA Ends (5′RACE) analysis, suggesting that vsiRNA-mediated host mRNA cleavage may not be widespread.
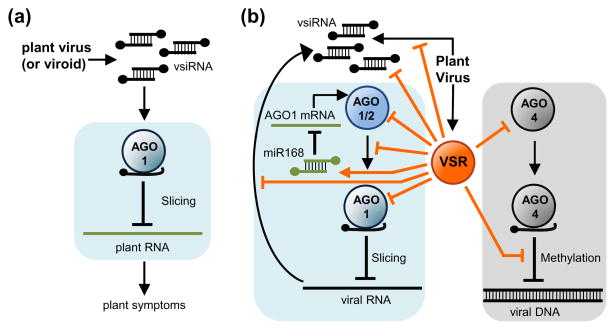
Figure 2.
Viral strategies to modulate or suppress plant RNA silencing. (a) VsiRNAs hijack plant AGOs to target sufficiently complementary host transcripts and induce symptoms. (b) Virus encoded VSRs suppress AGO functions at multiple levels. VSRs can impede AGO association with vsiRNAs, promote AGO degradation, down-regulate AGO1 levels by interfering with its homeostasis or inactivate programmed AGO/vsiRNA complexes. Light blue and grey boxes include steps unique to PTGS or TGS, respectively.
Viral suppressors interfere with AGO antiviral functions
VSRs can target virtually any step of the antiviral RNA silencing pathway (for a recent review see Ref. [5•]). AGOs are preferred targets of VSRs at multiple levels (see below and Figure 2b).
First, VSRs can prevent AGO association with vsiRNAs i) by blocking vsiRNA biogenesis through viral dsRNA binding (e.g. Tomato spotted wilt virus NSs) [33], by blocking DCL action (e.g. Rice yellow mottle virus P1) [34], by interacting or competing with components of the siRNA-generating pathways such as RDR6 (e.g. Rice yellow stunt rhabdovirus P6) [35] or SUPPRESSOR OF GENE SILENCING3 (SGS3) (e.g. Plantago asiatica mosaic virus TGBp1) [36], or by impairing vsiRNA stability (e.g. Sweet potato stunt crinivirus RNAseIII) [37]; ii) through direct binding and sequestration of vsiRNA duplexes away from antiviral AGOs as shown for multiple VSRs encoded by viruses of diverse genera (e.g. tombusviral P19, potyviral HC-Pro) [5,11]; iii) by promoting AGO degradation as observed for polerovirus and enamovirus. P0 proteins that induce AGO1 uridylation and subsequent degradation in a proteasome-independent manner [38–41] through the autophagy pathway [42], and for PVX P25 that activates AGO1 degradation through the proteasome pathway [43]; and iv) by physically interacting with AGOs through GW repetitive motifs, as shown by TCV-P38 interaction with unloaded Arabidopsis AGO1 [44] or AGO2 [15].
Second, VSRs can also block vsiRNA-programmed AGOs. It was first proposed that CMV 2b inhibits RISC activity via physical interaction with Arabidopsis AGO1 PAZ domain [45]. It was also shown that the in vitro suppression of AGO1 and AGO4 slicing activities by CMV 2b requires its physical interaction with AGOs, although this interaction was dispensable for RNA silencing suppression by CMV 2b [46]. Similarly, P1 from Sweet potato mild mottle virus was shown to inhibit RISC activity by binding loaded AGO1 through its GW motifs [47]. The importance of the Glycine-Tryptophan (GW) motifs in AGO1 binding and suppression activity was further demonstrated when Sweet potato feathery mottle virus P1 was converted into a VSR by including two additional WG/GW motifs [48]. Interestingly, it was recently reported that the inhibition of Pelargonium line pattern virus P37 suppressor activity caused by mutations in P37 W residues was most likely not due, at least uniquely, to the loss of P37 interaction with AGO1 and AGO4 [49•]. Indeed, mutations in the multifunctional P37 caused multiple concomitant effects, and it was proposed that the loss of P37 ability for binding vsiRNAs was actually the main reason explaining the loss of P37 suppression activity [49•]. VSRs also suppress AGO4-mediated TGS of viral DNAs as reported for geminiviral AL2 and L2 [50], AC2 [51] and C2 [52] proteins.
And third, VSRs can alleviate AGO1 antiviral function by interfering with its homeostatic regulatory loop. It was proposed that a common effect upon plant virus infections is the increase and decrease in the expression of AGO1 mRNA and AGO1 protein, respectively [53]. This apparent paradox could be explained by the translational repression of AGO1 mRNA by miR168, a miRNA ubiquitously induced in plant-virus infections and dependent on P19 VSR, at least in Cymbidum ringspot virus-infected plants [53]. More recently, it was confirmed that various unrelated VSRs enhance MIR168 transcription and subsequent reduction in AGO1 accumulation [54].
Conclusion
Despite the considerable efforts made in recent years towards understanding AGO antiviral functions in plants, several outstanding questions remain to be answered. What are the bases explaining the antiviral functional specialization of certain, but not all, AGOs? In the context of biological networks, what are the molecular signals that dictate the coordinated and cooperative antiviral activities of AGOs in different plant developmental stages and tissues? For RNA viruses, which viral RNAs do AGOs target preferentially – those undergoing replication or translation, or both? Which are the authentic AGO target sites in viral RNAs, and how exactly have AGOs evolved to target those specific sites and not others? In which subcellular compartment(s) does slicing or translational repression of viral RNAs occur? Given the broad interest of these fundamental questions, we anticipate that some of them will be answered soon.
AGO1 and AGO2 are the two major plant antiviral ARGONAUTES against RNA viruses
AGOs bind virus-derived small RNAs to target complementary viral RNA or DNA
Virus-derived small RNAs can hijack AGOs to target host transcripts
AGOs loaded with host small RNAs can regulate gene expression and promote defense
Virus-encoded silencing suppressors attenuate or block AGO antiviral activities
Acknowledgments
We thank members of the Carrington lab for useful and critical discussions, and apologize to those colleagues whose work could not be cited because of space and reference limitations. This work was supported by grants from the National Science Foundation (MCB-1231726, MCB-1330562) and National Institutes of Health (AI043288) to James C. Carrington, and from the European Commission (H2020-MSCA-IF-2014-655841) to Alberto Carbonell.
Footnotes
Conflict of interest
There is no conflict of interest relating to this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Alberto Carbonell, Email: acarbonell@ibmcp.upv.es.
James C. Carrington, Email: jcarrington@danforthcenter.org.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 2.Poulsen C, Vaucheret H, Brodersen P. Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures. Plant Cell. 2013;25:22–37. doi: 10.1105/tpc.112.105643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez de Alba AE, Elvira-Matelot E, Vaucheret H. Gene silencing in plants: A diversity of pathways. Biochim Biophys Acta. 2013;1829:1300–1308. doi: 10.1016/j.bbagrm.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Szittya G, Burgyan J. RNA interference-mediated intrinsic antiviral immunity in plants. Curr Top Microbiol Immunol. 2013;371:153–181. doi: 10.1007/978-3-642-37765-5_6. [DOI] [PubMed] [Google Scholar]
- 5•.Csorba T, Kontra L, Burgyan J. viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology. 2015;479–480:85–103. doi: 10.1016/j.virol.2015.02.028. This interesting review provides an exhaustive compilation of information related to the mechanistic functions of all VSRs identified to date in plant viruses (of special interest are Table 1 and Figure 1) [DOI] [PubMed] [Google Scholar]
- 6.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu F, Ye X, Morris TJ. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci U S A. 2008;105:14732–14737. doi: 10.1073/pnas.0805760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative Argonautes in Arabidopsis thaliana. Plant Cell. 2011;23:1625–1638. doi: 10.1105/tpc.110.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dzianott A, Sztuba-Solinska J, Bujarski JJ. Mutations in the antiviral RNAi defense pathway modify Brome mosaic virus RNA recombinant profiles. Mol Plant Microbe Interact. 2012;25:97–106. doi: 10.1094/MPMI-05-11-0137. [DOI] [PubMed] [Google Scholar]
- 11•.Garcia-Ruiz H, Carbonell A, Hoyer JS, Fahlgren N, Gilbert KB, Takeda A, Giampetruzzi A, Garcia Ruiz MT, McGinn MG, Lowery N, et al. Roles and Programming of Arabidopsis ARGONAUTE Proteins during Turnip Mosaic Virus Infection. PLoS Pathog. 2015;11:e1004755. doi: 10.1371/journal.ppat.1004755. In this work, a modular anti-TuMV activity of Arabidopsis AGO1, AGO2, AGO5, AGO7 and AGO10 in various organs was reported. In leaves, AGO2 had a major role, and in inflorescences AGO1 and AGO10 had overlapping antiviral functions after systemic movement of the virus. Interestingly, antiviral AGOs associated with vsiRNAs but only in the absence of HC-Pro, which sequesters vsiRNAs away from AGOs to suppress their antiviral functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harvey JJ, Lewsey MG, Patel K, Westwood J, Heimstadt S, Carr JP, Baulcombe DC. An antiviral defense role of AGO2 in plants. PLoS One. 2011;6:e14639. doi: 10.1371/journal.pone.0014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaubert MJ, Bhattacharjee S, Mello AF, Perry KL, Moffett P. AGO2 mediates RNA silencing anti-viral defenses against Potato virus X in Arabidopsis. Plant Physiol. 2011;156:1556–1564. doi: 10.1104/pp.111.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbonell A, Fahlgren N, Garcia-Ruiz H, Gilbert KB, Montgomery TA, Nguyen T, Cuperus JT, Carrington JC. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell. 2012;24:3613–3629. doi: 10.1105/tpc.112.099945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Zhang X, Singh J, Li D, Qu F. Temperature-dependent survival of Turnip crinkle virus-infected arabidopsis plants relies on an RNA silencing-based defense that requires dcl2, AGO2, and HEN1. J Virol. 2012;86:6847–6854. doi: 10.1128/JVI.00497-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma X, Nicole MC, Meteignier LV, Hong N, Wang G, Moffett P. Different roles for RNA silencing and RNA processing components in virus recovery and virus-induced gene silencing in plants. J Exp Bot. 2015;66:919–932. doi: 10.1093/jxb/eru447. [DOI] [PubMed] [Google Scholar]
- 17.Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 18.Hamera S, Song X, Su L, Chen X, Fang R. Cucumber mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities. Plant J. 2012;69:104–115. doi: 10.1111/j.1365-313X.2011.04774.x. [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharjee S, Zamora A, Azhar MT, Sacco MA, Lambert LH, Moffett P. Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J. 2009;58:940–951. doi: 10.1111/j.1365-313X.2009.03832.x. [DOI] [PubMed] [Google Scholar]
- 20.Raja P, Sanville BC, Buchmann RC, Bisaro DM. Viral genome methylation as an epigenetic defense against geminiviruses. J Virol. 2008;82:8997–9007. doi: 10.1128/JVI.00719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raja P, Jackel JN, Li S, Heard IM, Bisaro DM. Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J Virol. 2014;88:2611–2622. doi: 10.1128/JVI.02305-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scholthof HB, Alvarado VY, Vega-Arreguin JC, Ciomperlik J, Odokonyero D, Brosseau C, Jaubert M, Zamora A, Moffett P. Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana. Plant Physiol. 2011;156:1548–1555. doi: 10.1104/pp.111.178764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghoshal B, Sanfacon H. Temperature-dependent symptom recovery in Nicotiana benthamiana plants infected with tomato ringspot virus is associated with reduced translation of viral RNA2 and requires ARGONAUTE 1. Virology. 2014;456–457:188–197. doi: 10.1016/j.virol.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 24••.Wu J, Yang Z, Wang Y, Zheng L, Ye R, Ji Y, Zhao S, Ji S, Liu R, Xu L, et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. Elife. 2015;4 doi: 10.7554/eLife.05733. In this work, the authors describe the cooperative antiviral action of rice AGO1 and AGO18. AGO18 is induced upon viral infection, and sequesters miR168 away from AGO1 thus disrupting AGO1 homeostasis, increasing AGO1 accumulation and enhancing its antiviral activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell. 2010;39:282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 26•.Schuck J, Gursinsky T, Pantaleo V, Burgyan J, Behrens SE. AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Res. 2013;41:5090–5103. doi: 10.1093/nar/gkt193. The authors use cytoplasmic extracts of evacuolated tobacco protoplasts to test the AGO-mediated in vitro cleaveage of replicating viral RNAs. They show that RISCs containing AGO1, AGO2, AGO3 and AGO5 loaded synthetic vsiRNAs and inhibited viral replication, particularly AGO1 and AGO2. These two AGOs targeted neighboring sequences in TBSV RNA suggesting that some regions of the viral RNAs could be more accessible and collectively targeted by multiple AGOs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H, Duan CG, Hou WN, Du QS, Lv DQ, Fang RX, Guo HS. Satellite RNA-derived small interfering RNA satsiR-12 targeting the 3′ untranslated region of Cucumber mosaic virus triggers viral RNAs for degradation. J Virol. 2011;85:13384–13397. doi: 10.1128/JVI.05806-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Cao M, Du P, Wang X, Yu YQ, Qiu YH, Li W, Gal-On A, Zhou C, Li Y, Ding SW. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc Natl Acad Sci U S A. 2014;111:14613–14618. doi: 10.1073/pnas.1407131111. This interesting work identifies a new class of Arabidopsis endogenous siRNAs, named vasiRNAs, that are activated upon virus infection. VasiRNAs are DRB1/RDR1-dependent and function through AGO2 to target multiple host genes and promote antiviral defense. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith NA, Eamens AL, Wang MB. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 2011;7:e1002022. doi: 10.1371/journal.ppat.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba J, Sueda K, Burgyan J, Masuta C. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 2011;7:e1002021. doi: 10.1371/journal.ppat.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navarro B, Gisel A, Rodio ME, Delgado S, Flores R, Di Serio F. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012;70:991–1003. doi: 10.1111/j.1365-313X.2012.04940.x. [DOI] [PubMed] [Google Scholar]
- 32.Miozzi L, Gambino G, Burgyan J, Pantaleo V. Genome-wide identification of viral and host transcripts targeted by viral siRNAs in Vitis vinifera. Mol Plant Pathol. 2013;14:30–43. doi: 10.1111/j.1364-3703.2012.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Ronde D, Pasquier A, Ying S, Butterbach P, Lohuis D, Kormelink R. Analysis of Tomato spotted wilt virus NSs protein indicates the importance of the N-terminal domain for avirulence and RNA silencing suppression. Mol Plant Pathol. 2014;15:185–195. doi: 10.1111/mpp.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lacombe S, Bangratz M, Vignols F, Brugidou C. The rice yellow mottle virus P1 protein exhibits dual functions to suppress and activate gene silencing. Plant J. 2010 doi: 10.1111/j.1365-313X.2009.04062.x. [DOI] [PubMed] [Google Scholar]
- 35.Guo H, Song X, Xie C, Huo Y, Zhang F, Chen X, Geng Y, Fang R. Rice yellow stunt rhabdovirus protein 6 suppresses systemic RNA silencing by blocking RDR6-mediated secondary siRNA synthesis. Mol Plant Microbe Interact. 2013;26:927–936. doi: 10.1094/MPMI-02-13-0040-R. [DOI] [PubMed] [Google Scholar]
- 36.Okano Y, Senshu H, Hashimoto M, Neriya Y, Netsu O, Minato N, Yoshida T, Maejima K, Oshima K, Komatsu K, et al. In Planta Recognition of a Double-Stranded RNA Synthesis Protein Complex by a Potexviral RNA Silencing Suppressor. Plant Cell. 2014;26:2168–2183. doi: 10.1105/tpc.113.120535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinheimer I, Jiu Y, Rajamaki ML, Matilainen O, Kallijarvi J, Cuellar WJ, Lu R, Saarma M, Holmberg CI, Jantti J, et al. Suppression of RNAi by dsRNA-Degrading RNaseIII Enzymes of Viruses in Animals and Plants. PLoS Pathog. 2015;11:e1004711. doi: 10.1371/journal.ppat.1004711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol. 2007;17:1609–1614. doi: 10.1016/j.cub.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 39.Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P, Ziegler-Graff V. The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr Biol. 2007;17:1615–1621. doi: 10.1016/j.cub.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 40.Csorba T, Lozsa R, Hutvagner G, Burgyan J. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 2010;62:463–472. doi: 10.1111/j.1365-313X.2010.04163.x. [DOI] [PubMed] [Google Scholar]
- 41.Fusaro AF, Correa RL, Nakasugi K, Jackson C, Kawchuk L, Vaslin MF, Waterhouse PM. The Enamovirus P0 protein is a silencing suppressor which inhibits local and systemic RNA silencing through AGO1 degradation. Virology. 2012;426:178–187. doi: 10.1016/j.virol.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 42.Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, Genschik P. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci USA. 2012;109:15942–15946. doi: 10.1073/pnas.1209487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiu MH, Chen IH, Baulcombe DC, Tsai CH. The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol Plant Pathol. 2010;11:641–649. doi: 10.1111/j.1364-3703.2010.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, Garcia S, Braun L, Bergdoll M, Hakimi MA, Lagrange T, et al. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 2010;24:904–915. doi: 10.1101/gad.1908710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duan CG, Fang YY, Zhou BJ, Zhao JH, Hou WN, Zhu H, Ding SW, Guo HS. Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell. 2012;24:259–274. doi: 10.1105/tpc.111.092718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giner A, Lakatos L, Garcia-Chapa M, Lopez-Moya JJ, Burgyan J. Viral Protein Inhibits RISC Activity by Argonaute Binding through Conserved WG/GW Motifs. PLoS Pathog. 2010;6:e1000996. doi: 10.1371/journal.ppat.1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szabo EZ, Manczinger M, Goblos A, Kemeny L, Lakatos L. Switching on RNA silencing suppressor activity by restoring argonaute binding to a viral protein. J Virol. 2012;86:8324–8327. doi: 10.1128/JVI.00627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49•.Perez-Canamas M, Hernandez C. Key Importance of Small RNA Binding for the Activity of a Glycine-Tryptophan (GW) Motif-containing Viral Suppressor of RNA Silencing. J Biol Chem. 2015;290:3106–3120. doi: 10.1074/jbc.M114.593707. This paper reports that the mutation of W residues in P37 (the VSR of PLPV) suppresses its interaction with AGOs and its silencing suppression activity. However, further analyses show that indeed the mutations in P37 leed to other concomitant effects and affect other P37 functions. Indeed, vsiRNA binding seems the main suppression mechanism of P37, although the cooperation of multiple suppression mechanisms including AGO-binding is not discarded. The authors suggest to not overlook possible concomitant effects when inactivating multifunctional VSRs that could lead to misinterpretations of results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J Virol. 2009;83:5005–5013. doi: 10.1128/JVI.01771-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soitamo AJ, Jada B, Lehto K. Expression of geminiviral AC2 RNA silencing suppressor changes sugar and jasmonate responsive gene expression in transgenic tobacco plants. BMC Plant Biol. 2012;12:204. doi: 10.1186/1471-2229-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, Chen H, Huang X, Xia R, Zhao Q, Lai J, Teng K, Li Y, Liang L, Du Q, et al. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell. 2011;23:273–288. doi: 10.1105/tpc.110.081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varallyay E, Valoczi A, Agyi A, Burgyan J, Havelda Z. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 2010;29:3507–3519. doi: 10.1038/emboj.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varallyay E, Havelda Z. Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Mol Plant Pathol. 2013;14:567–575. doi: 10.1111/mpp.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]