Fig. 1.
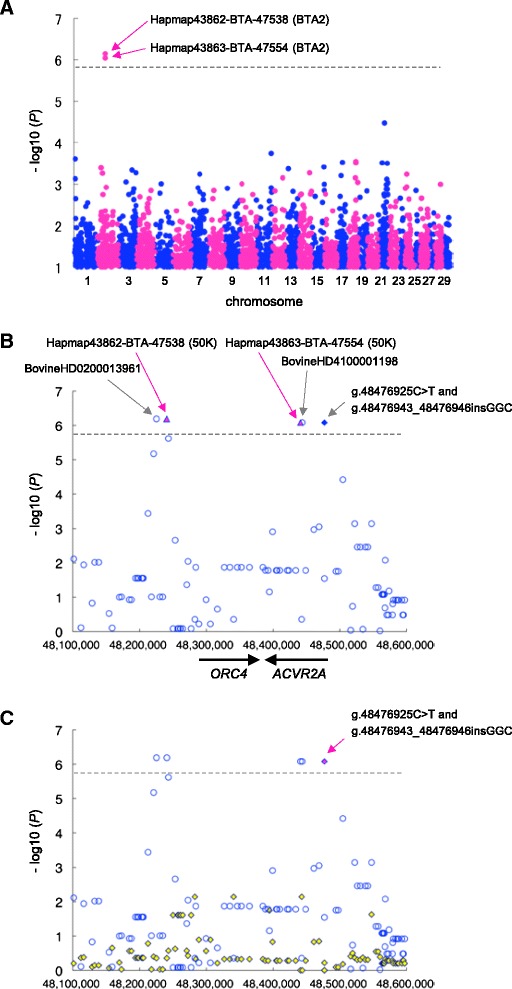
Association of SNPs and a indel with ANAI4 in 430 Japanese Black cattle. a Manhattan plot of the association of 33,303 SNPs (BovineSNP50K BeadChip) with ANAI4 in 430 Japanese Black cattle. The chromosomes are distinguished with alternating colors (blue, odd numbers; red, even numbers). The chromosome number is indicated on the X-axis. The dashed line is the Bonferroni-corrected threshold for genome-wide significance (−log10 (P) = 5.823). The vertical axis is broken for P values below -log10 (P) = 1. b Regional plot of the locus on BTA2 associated with ANAI4. SNPs from the BovineSNP50K BeadChip are shown as red triangles. The imputed SNPs are shown as unfilled blue circles. g.48476925C > T SNP and the g.48476943_48476946insGGC indel are shown as filled blue diamonds. Genes and their directions of transcription are noted at the bottom of the plot. c A conditioned analysis was performed by including the haplotype, defined by g.48476925C > T SNP and the g.48476943_48476946insGGC indel, as covariates in the model. The 2 red, filled diamonds indicate the g.48476925C > T and g.48476943_48476946insGGC (arrow) variants. The blue, unfilled circles and the yellow, filled diamonds represent P values on a –log10 scale before and after conditioning, respectively. The positions shown are based on the UMD3.1 assembly of the bovine genome
