Fig 6.
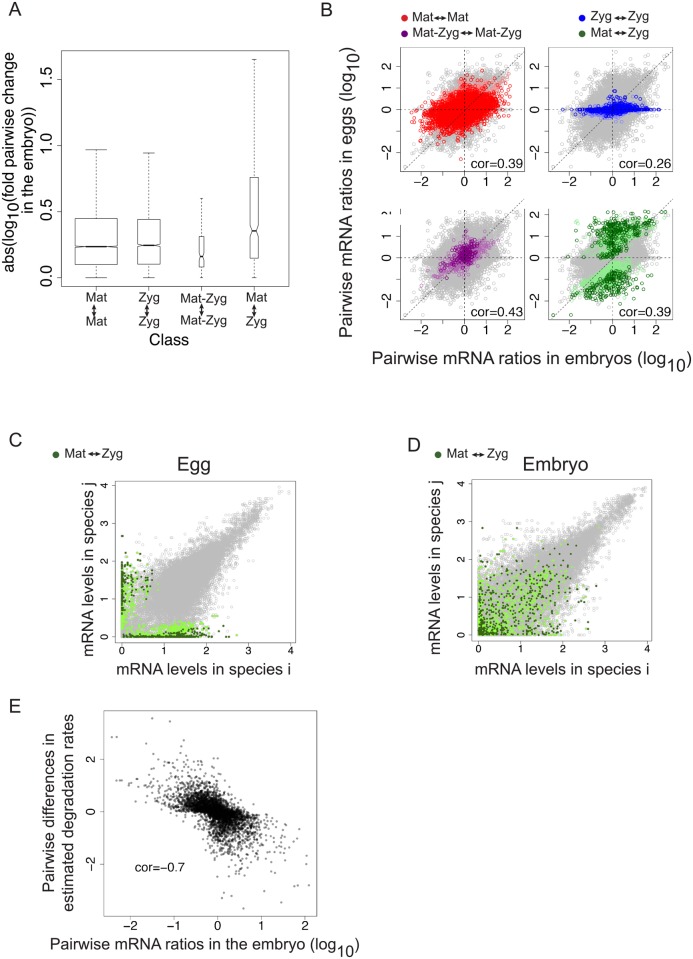
A. Genes whose origin (“Mat <-> Zyg”) changes show higher species differences in embryonic mRNA levels compared to genes with conserved origin. B. Divergence of mRNA levels between species is correlated with divergence of mRNA levels in the egg. We compared pairwise species differences of mRNA levels in the embryo and in the egg and found an overall positive correlation. We focused on the genes that had a conserved maternal (red), zygotic (blue) or Mat-Zyg (purple) origin in two species, or a change from maternal to zygotic (green). Grey points represent the complete dataset (all pairwise comparisons for the 5985 genes). C/D. Genes that have a divergent maternal or zygotic origin (“Mat <-> Zyg”) show highly divergent mRNA levels in the egg (C), which partially translates in the embryo (B). Points in green represent genes that are classified as maternal or zygotic in one species and have switched to zygotic or maternal in the other. Dark green was used for genes classified with high probabilities and light green was used for genes classified with low probabilities. E. Divergence of degradation rates correlates with embryonic expression in the embryo: an increase in mRNAs between species correlates with a decrease in degradation rates. We compared species differences in estimated degradation rates with species differences in mRNA levels in the embryo for the 667 genes that were predicted to have species-specific degradation rates according to a likelihood ratio test. Spearman correlation rho = 0.7, p. value < 2.2x10-16.