Figure 5.
Tbx3 Represses Wnt Signaling-Mediated Mesoderm Differentiation
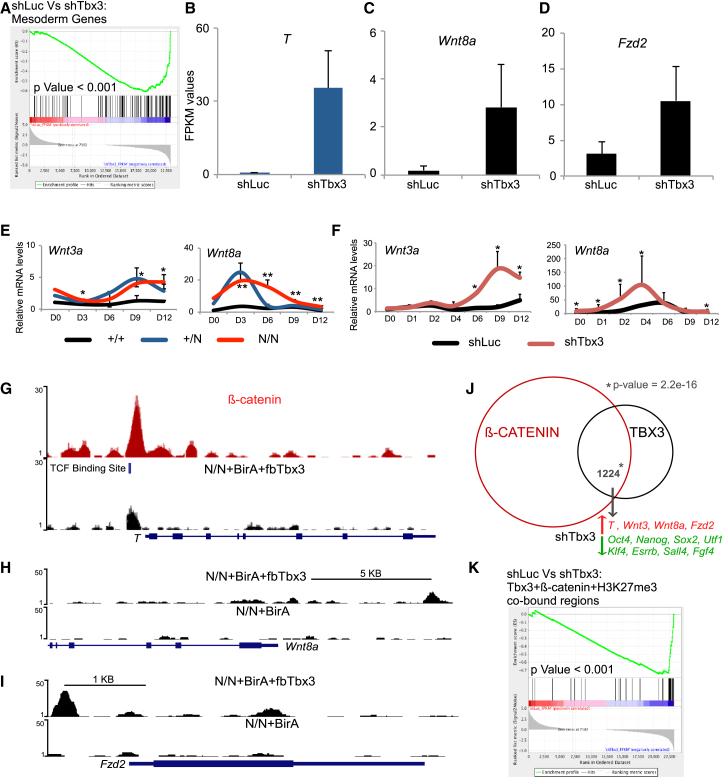
(A) Gene set enrichment analysis (GSEA) to detect statistical significance of mesoderm genes in shTbx3 versus shLuc. Cells grown in ESC media containing LIF were infected with lentiviral shRNA constructs against Luciferase (shLuc) or Tbx3 (shTbx3).
(B–D) mRNA-Seq FPKM values of T, Wnt8a, and Fzd2 in cells infected the indicated shRNAs.
(E) mRNA expression profile of indicated genes during EB differentiation time course of 12 days for indicated cell lines. Data are the average of three independent replicates. ∗p < 0.05 and ∗∗p < 0.005 (t test).
(F) mRNA expression profile of the indicated genes during EB differentiation time course of 12 days in WT mESCs infected with indicated shRNAs. Data are the average of three independent replicates. ∗p < 0.05 and ∗∗p < 0.005 (t test).
(G–I) TBX3 ChIP-seq binding data from N/N-BirA (control) and N/N-BirA+fbTbx3 cells. β-CATENIN binding from published data.
(J) Venn diagram representing overlap between β-CATENIN and TBX3 genome-wide binding sites. The p value was generated using the fisher exact test.
(K) GSEA to detect statistical significance of TBX3, β-CATENIN, and H3K27me3 co-bound gene set in shTbx3 versus shLuc.