Figure 1.
H3K4 Trimethylation Is Cell-Cycle Regulated at Bivalent Domains of Developmentally Regulated Genes
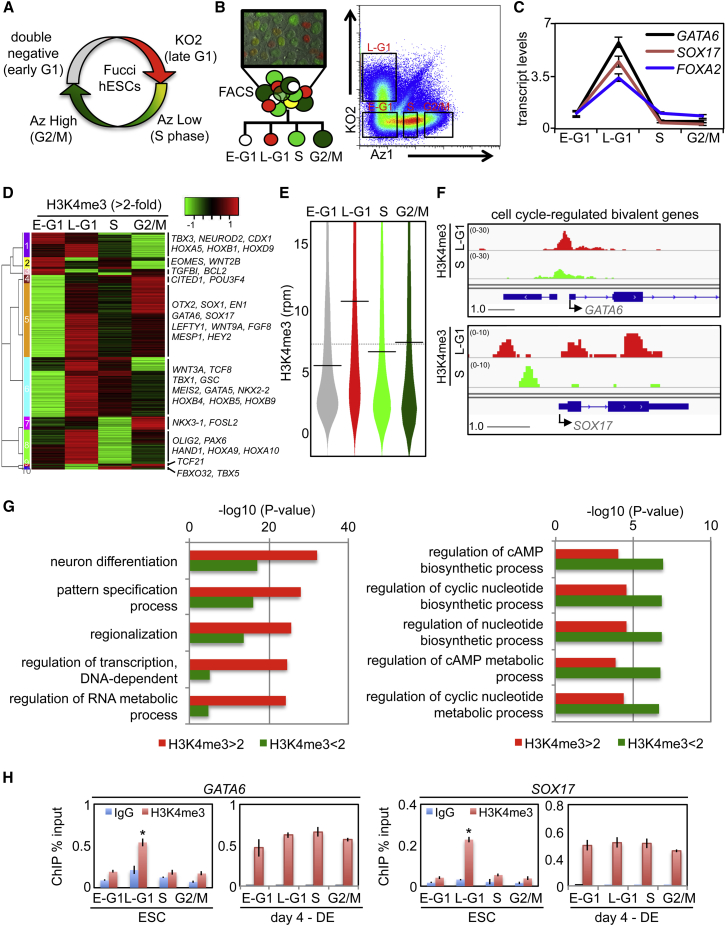
(A) Diagram of the Fucci reporter system established in WA09 hESCs.
(B) The Fucci system can be used to isolate cell-cycle fractions in live hESC cells by FACS.
(C) qRT-PCR of Fucci-sorted cells exhibit periodicity of transcription for developmental genes. Data are the average of technical triplicates and representative of more than ten independent experiments.
(D) ChIP-seq of bivalent genes with cell-cycle-regulated H3K4me3 (>2-fold change between any two fractions) represented as a heatmap. Representative, developmental genes belonging to different cell-cycle-regulated clusters are shown. ChIP-seq data are from the pool of three independent experiments.
(E) Bean-plot diagram showing distribution of H3K4me3 levels for cell-cycle-regulated bivalent genes. The black horizontal line represents median values. Late G1 was significantly higher than S (p < 1 × 10−14). Statistical analyses are described in ChIP-seq data and are from the pool of three independent experiments.
(F) H3K4me3 ChIP-seq profiles of hESC Fucci fractions for GATA6 and SOX17. Scale bar indicates genomic distance in kb and value range given in RPM. ChIP-seq data are from the pool of three independent experiments.
(G) GO analysis comparing cell-cycle-regulated bivalent genes and non-cell-cycle-regulated bivalent genes.
(H) H3K4me3 qChIP using Fucci-isolated cell-cycle fractions from ESCs or DE at the GATA6 or SOX17 promoters. Data are the average of three independent replicates.
∗p < 0.05. Error bars in this figure represent the SEM. See also Figures S1 and S2 and Tables S1 and S2.