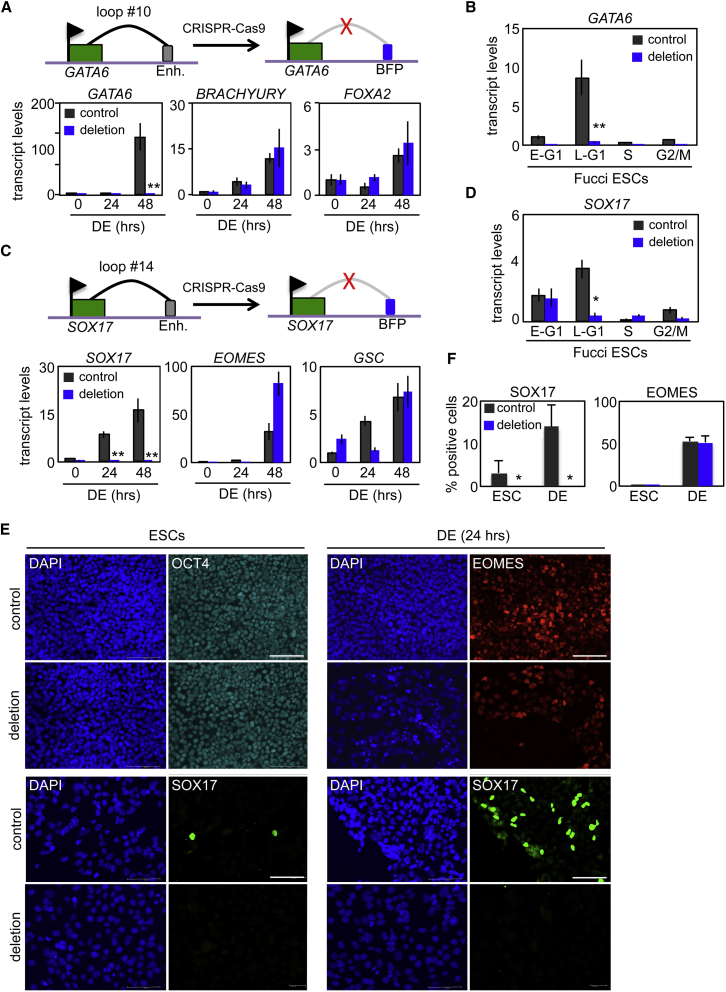
Figure 5.
Cell-Cycle-Dependent Recruitment of Enhancer Elements Are Required for the Activation of Bivalent Genes
(A) Organization of the GATA6 locus around a region displaying cell-cycle-dependent changes in chromosome architecture. The specific loop structure (loop #10), its associated enhancer region, and CRISPR-Cas9-directed disruption of a 1 kb region encompassing the enhancer and the resulting insertion of a BFP reporter cassette are indicated (top). qRT-PCR transcript analysis of WT (control) and genome-edited (deletion) ESCs was performed over the first 48 hr of differentiation toward DE for indicated genes (bottom).
(B) Analysis of GATA6 transcript levels during the cell cycle in Fucci ESC cell-cycle fractions from WT (control) and genome-edited (deletion; loop #10) cells. Data are the average of three independent replicates.
(C) Organization of the SOX17 locus around a region displaying cell-cycle-dependent changes in chromosome architecture. The specific lop structure (loop #14), its associated enhancer region, and CRISPR-Cas9-directed disruption of a 1 kb region encompassing the enhancer and the resulting insertion of a BFP reporter cassette are indicated (top). qRT-PCR transcript analysis of WT (control) and genome-edited (deletion) ESCs was performed over the first 48 hr of differentiation toward DE for indicated genes (bottom). Data are the average of three independent replicates.
(D) Analysis of SOX17 transcript levels during the cell cycle in Fucci ESC cell-cycle fractions from WT (control) and genome-edited (deletion; loop #14) cells. Data are the average of three independent replicates.
(E) Immunostaining of WT (control) and genome-edited (SOX17 locus; loop #14) cells under self-renewal (ESCs) or differentiation (DE, 24 hr) conditions.
(F) Quantitation of immunostaining in (E) from three separate fields of view for each antibody (n > 1000). Micron bar is 100 μm. Data are representative of three independent experiments.
∗p < 0.05, ∗∗p < 0.01. Error bars in this figure represent the SEM. See also Figure S5.