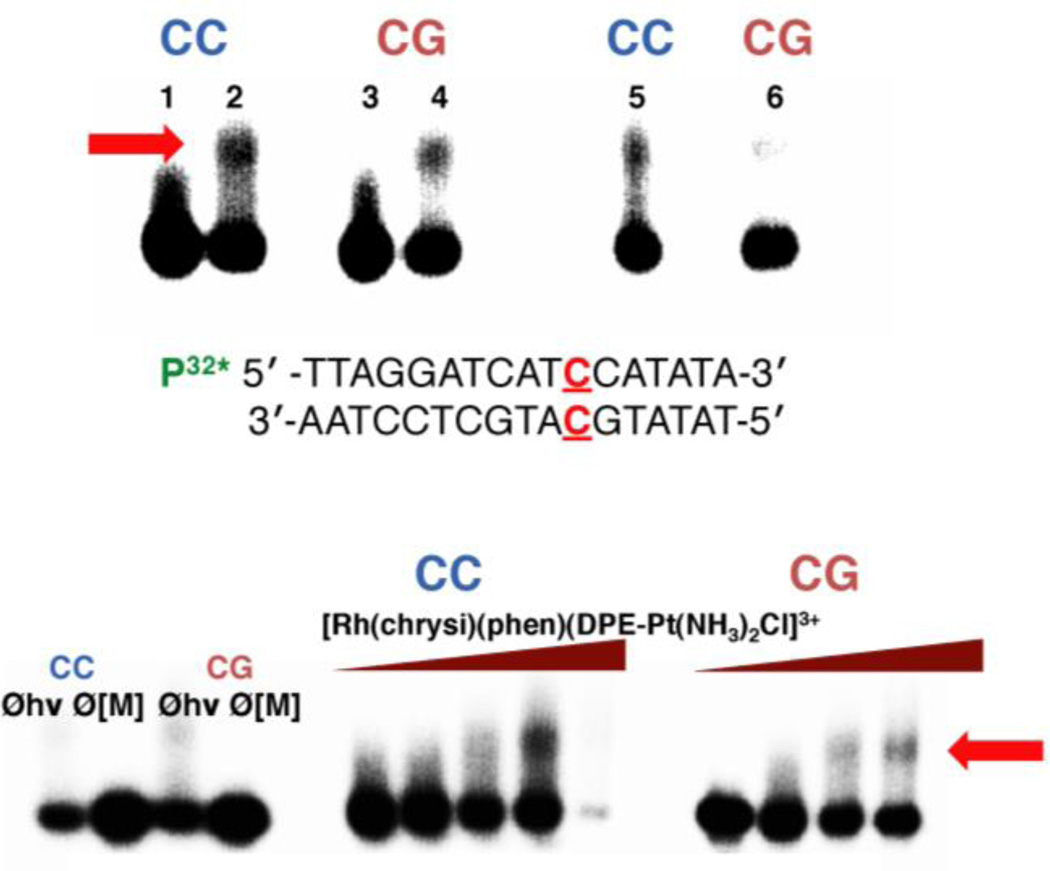
Figure 3.
Platinum binding to mismatched and well-matched DNA. Samples were irradiated for 15 min and electrophoresed on a 20% denaturing PAGE gel. Platinum crosslinking of 5’-32P-labelled DNA is indicated by the appearance of slow-moving bands located above the unmodified parent DNA; platinated DNA is denoted by a red arrow. (Top) Autoradiogram showing the formation of covalent platinum adducts with mismatched and well-matched DNA duplexes (1 µM ) as a function of time. [Rh(chrysi)(phen)(DPE-Pt(NH3)2Cl)]3+ (1 µM ) was incubated with 5’end radiolabeled duplex DNA of the sequence indicated (bottom; the site of the CC mismatch is denoted in red) as well as the corresponding well-matched duplex in buffer (75 mM NaCl, 10 mM NaPi, pH 7.1) at 37 °C for 1, 3, or 18h (left to right). Lanes: (1) untreated duplex DNA containing a CC mismatch; (2) mismatched DNA incubated with metal complex for 1h; (3) untreated well-matched DNA; (4) well-matched DNA incubated with metal complex for 1h; (5) mismatched DNA treated with metal complex for 3h; (6) well-matched DNA treated with metal complex for 3h. Samples treated with metal complex for 18h were degraded on the gel and are not visible in the autoradiogram. (Bottom) Autoradiogram showing the formation of covalent platinum adducts with mismatched and well-matched DNA duplexes (1 µM ) as a function of metalloinsertor concentration. Controls without irradiation (Øhν) and without metal complex (Ø[M]) were included for each type of DNA (mismatched DNA is denoted by “CC” in blue; well-matched DNA is denoted by “CG” in red) and are depicted on the left.