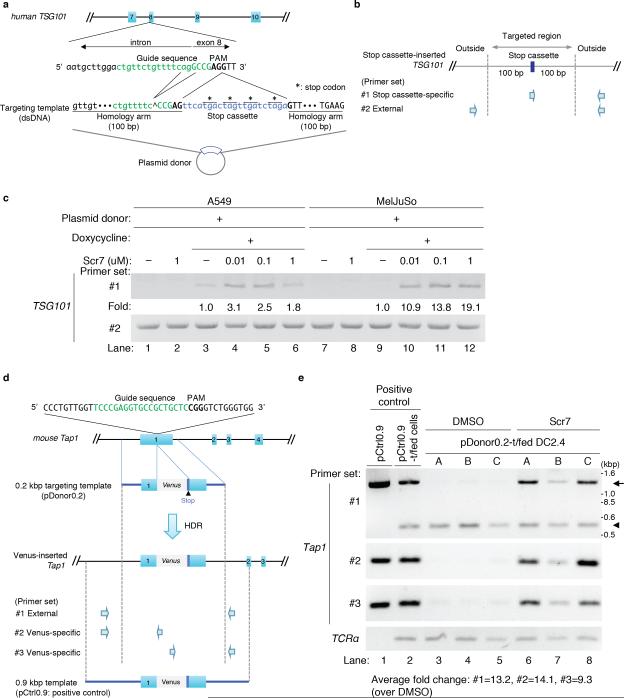
Figure 1. The NHEJ inhibitor Scr7 enhances the efficiency of insertional mutagenesis in cell lines.
(a) Schematic of sgRNA/ targeting template-targeting site at TSG101 exon 8. The guide sequence of human TSG101 sgRNA is indicated in green. The protospacer-adjacent motif (PAM) is in bold typeface. The exon sequence is capitalized. The targeting template contains 100 base pairs (bp) homology arms that the double strand breaks (DSBs) on both sides. To avoid CRISPR/Cas9-based DSBs in the plasmid donor containing the targeting template, the ‘agg’ nucleotides in the guide sequence were deleted. The stop cassette is labeled in blue. (*: stop codon).
(b) Primer sets to detect the insertion of the stop cassette. The presence of the stop cassette in the TSG101 locus was analyzed using an internal forward primer specific for the stop cassette and an external reverse primer specific for a sequence outside of the flanking arm (Fig. 1b primer set #1, reverse primer: 419 bp downstream of the PAM motif). The genomic region is targeted by external primers as an internal control (Fig. 1b, primer set #2, forward primer: 229 bp upstream of the PAM motif, reverse primer: identical to the reverse primer of set #1).
(c) Increased frequency of HDR in the presence of Scr7 in A549 and MelJuSo cells. The Cas9-inducible A549 and MelJuSo cell lines stably expressing TSG101 sgRNA were transfected with the plasmid donor. Twenty-four hours after transfection, cells were treated with doxycycline and Scr7 at the indicated concentration for another 24 hours. Doxycycline and Scr7 were removed by washing, and the cells were cultured for another 24 hours in fresh media, and finally lysed to collect genomic DNA. Analyses were performed using primer sets #1 and #2. Data are representative of 3 experiments. The HDR frequency -fold change was calculated by determining the ratio of band intensities at the different concentrations of Scr7 (lane 4-6 and lane 10-12) over the intensities observed in the absence of Scr7 (lane 3 and 9). See Online Methods for the detailed method for quantification of band intensities.
(d) Strategy for insertion of a Venus reporter gene into the mouse Tap1 exon1. The guide sequence of mouse Tap1 sgRNA is indicated in green. The PAM is designated by bold-typeface. The targeting template was introduced into a plasmid (plasmid donor). A Venus reporter gene (813 bp) is shown in gray and contains the stop codon. We prepared two constructs: pDonor0.2 as a template donor and pCtrl0.9 as a positive control for a PCR. pDonor0.2 contains 213 base pair (bp) flanking arms on either side of a Venus reporter gene to induce HDR in front of the PAM motif (GFP mutant Venus size: 813 bp); pCtrl0.9 has longer homology arms (Fig. 1f, bottom; the length of upstream homology arm: 914 bps; downstream length: 660 bp), and was used to provide a comparative PCR product of the Venus-inserted Tap1 locus in lane 1 and 2 of Figure 1g.
Primer sets to detect the insertion of the Venus gene and positive control plasmid. External primers were designed outside of the homology arms of the 200 bp targeting template. Primer set #1: a pair of external forward and reverse primers designed outside of the homology arms on the 0.2 kbp targeting template (forward primer: 258 bp upstream of the PAM motif, reverse primer: 252 bp downstream of the PAM motif), primer set #2: the forward primer from primer set #1 and a Venus-specific reverse primer, primer set #3: a Venus-specific forward primer and the external reverse primer from primer set #1. As a positive control for PCR, Vector pCtrl0.9 containing longer homology arms flanking both sides was prepared (length of the left homology arm: 914 bps, length of the right homology arm: 660 bp).
(e) Improvement of HDR-mediated insertion efficiency by inclusion of Scr7 in DC2.4 cells. Lane 1 serves as a positive control, using the pCtrl0.9 as a PCR template to provide the comparative PCR products of a Venus-inserted Tap1 locus. The Cas9-inducible DC2.4 cell line stably expressing Tap1 sgRNA was transfected with the pCtrl0.9 as an additional positive control (lane 2), or with pDonor0.2 (lane 3-8). After 24 hours of transfection, cells were treated with doxycycline and 1 μM of Scr7 for another 24 hours, after which Scr7 and doxycycline were removed by washing. The cells were cultured for another 24 hours, and lysed to collect genomic DNA. The genomic DNA was analyzed using the primer sets as shown in Fig. 1g. (arrow: the amplicon of the Venus-inserted Tap1 locus, arrowhead: the amplicon of original Tap1 locus) The samples analyzed were derived from 3 independent experiments (A-C), i.e. the sample in lane 3 is matched with the sample in lane 6, 4 with 5, and 7 with 8. The fold change in HDR frequency was calculated by determining the ratio of band intensities in the presence of Scr7 (lane 3-5) over the intensities observed in the absence of Scr7 (lane 6-8). For the panel of primer set #1, the ratio of the inserted band intensities (arrow) over total band intensities (arrow + arrowhead) was calculated, and then the -fold changes were determined. We observed the appearance of an ~800 bp larger DNA fragment (arrow) upon HDR-based Venus insertion as compared to the original Tap1 DNA fragment (arrowhead) (Fig. 1g, upper panel, individual experiment: A-C), as evidence for integration of the Venus gene into the target locus. Sequence analysis confirmed that the new, longer DNA fragment (Fig. 1g, bands indicated by arrow) amplified by primer set #1 corresponded to precise insertion of the Venus reporter gene (Supplementary Fig. 4a).