Fig. 3.
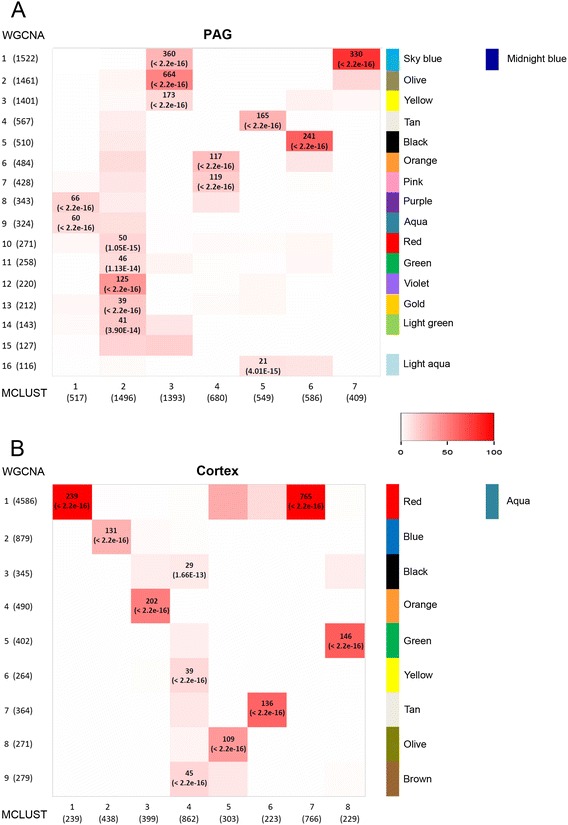
Conserved modules were identified by both the MCLUST and WGCNA methods. a Pairwise comparisons were made of the 16 WGCNA and 7 MCLUST modules identified in the PAG. 17 conserved modules were identified. All but one displayed a highly significant degree of overlap (p < 0.001). b Pairwise comparisons were made of the 9 WGCNA and 8 MCLUST modules identified in the cortex. 10 conserved modules were identified. All conserved modules displayed a highly significant degree of overlap (p < 0.001). The shading in the tables illustrates the percentage overlap as per the inset key. Significance of the most highly conserved modules for each pairwise comparison is given alongside the number of overlapping genes per conserved module. For simplicity we now assigned colour names to the conserved modules, which are given to the right of each corresponding row where the conserved module appears
