Fig. 1.
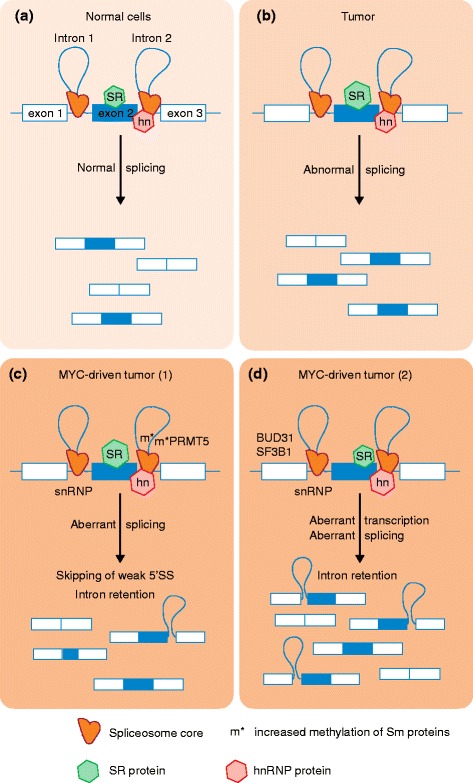
Splicing alterations in tumors. a In normal cells, the spliceosome, which is regulated by activators and repressors such as various serine-arginine-rich (SR) and heterogeneous nuclear ribonucleoprotein (hn) proteins, catalyzes pre-mRNA splicing, resulting in a normal, cell-type-specific splicing pattern. b In tumors, upregulation of certain splicing factors, for example SR proteins, or mutations in these factors promote abnormal splicing [3, 6, 7], leading to cancer-specific splicing patterns. c In the context of MYC-driven tumors, MYC directly upregulates transcription of splicing components, such as the splicing activator SR proteins and repressor hnRNP proteins [3, 6, 7], the PRMT5 methyltransferase, which controls Sm protein methylation [5], or the genes encoding snRNP constituents or snRNP assembly factors [5]. MYC-driven cancer cells exhibit aberrant splicing patterns, characterized by increased intron retention, and by increased skipping of exons that have weak 5′ splice sites (SS). d Alternatively, hyperactivation of MYC can lead to global upregulation of pre-mRNA levels, without directly affecting the expression of spliceosome components, and this excess of pre-mRNA overwhelms the splicing machinery [4]
