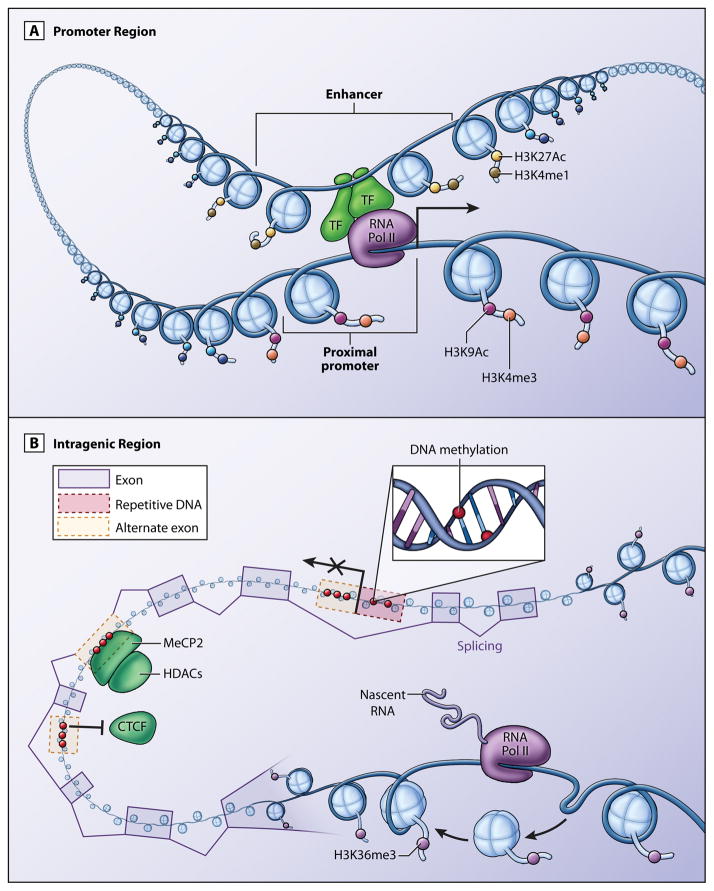
Figure 1.
Epigenetic-based mechanisms of gene regulation. (A) The epigenetic signatures at the promoter and enhancer of a gene contribute to the recruitment of trans factors (TF) and RNA Polymerase II (Pol II) for transcription initiation regulation. Epigenetic modifications that are observed frequently at proximal promoter regions include H3K9Ac (pink) and H3K4me3 (orange). Common epigenetic modifications that are associated with an active enhancer are H3K4me1 (olive) and H3K27Ac (yellow). (B) Intragenic epigenetic modifications are critical for transcription and mRNA variant expression. H3K36me3 (purple) is commonly associated with transcription elongation by RNA polymerase II through intragenic regions. HK36me3 is believed to be necessary for the inhibition of spurious transcription initiation. Intragenic DNA methylation (red) is positively correlated with gene expression and may prevent spurious transcript initiation from DNA repetitive elements or alternative promoters. Intragenic DNA methylation also affects alternative exon inclusion. DNA methylation may interfere with CTCF recruitment to inhibit exon inclusion. Alternatively, DNA methylation may recruit MeCP2 and HDACs to promote exon inclusion.