Abstract
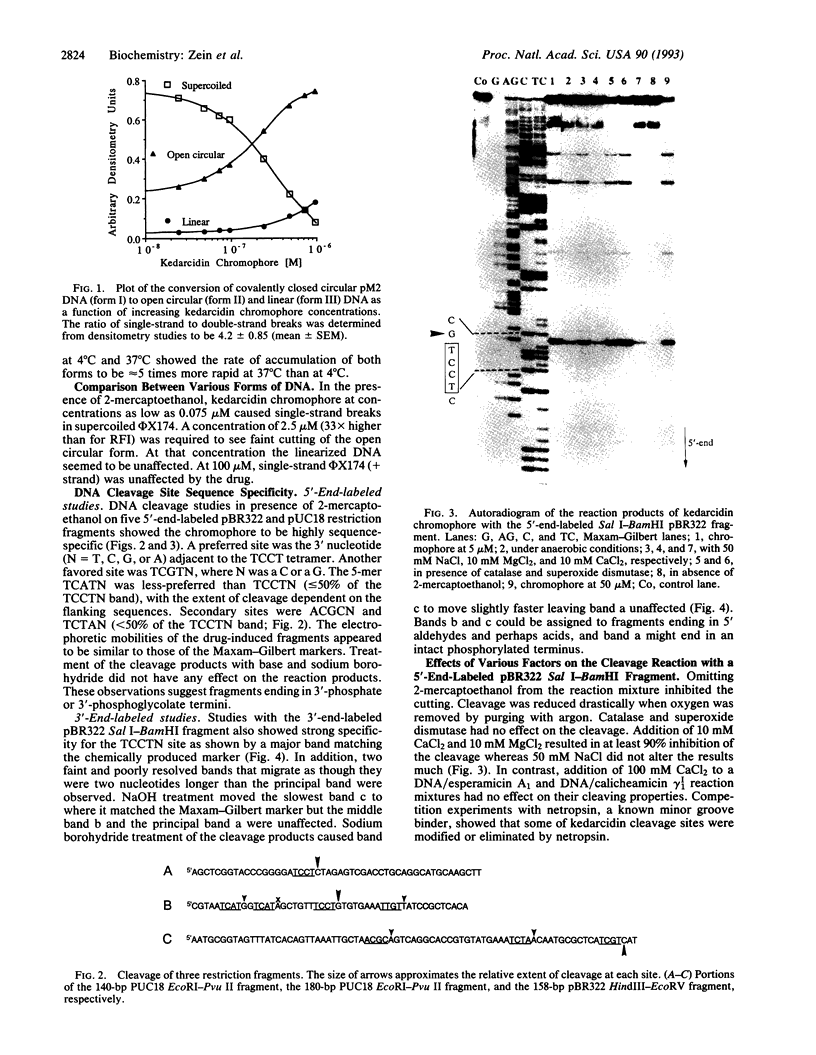
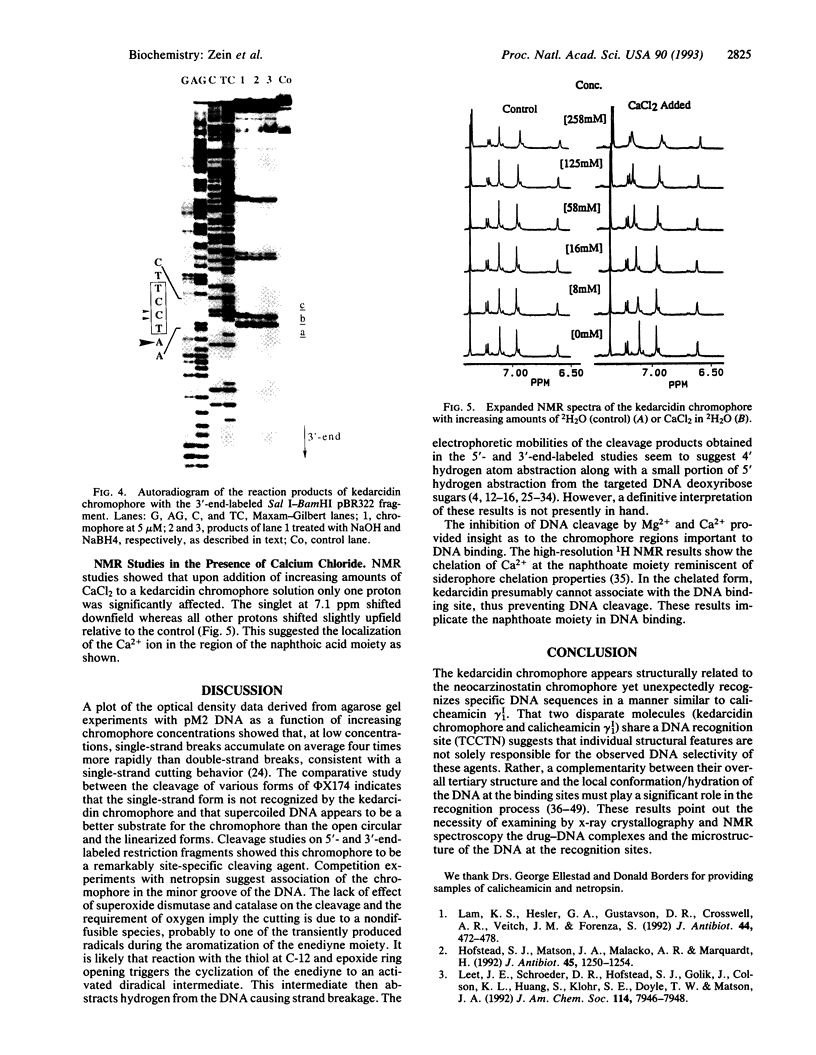
Kedarcidin chromophore is a 9-membered enediyne, recently isolated from an actinomycete strain. In vivo studies show this molecule to be extremely active against P388 leukemia and B16 melanoma. Cytotoxicity assays on the HCT116 colon carcinoma cell line result in an IC50 value of 1 nM. In vitro experiments with phi X174, pM2 DNA, and 32P-end-labeled restriction fragments demonstrate that this chromophore binds and cleaves duplex DNA with a remarkable sequence selectivity producing single-strand breaks. The cleavage chemistry requires reducing agents and oxygen similar to the other naturally occurring enediynes. Certain cations (Ca2+ and Mg2+) prevent strand cleavage. High-resolution 1H NMR studies on the chromophore in the presence of calcium chloride implicate the 2-hydroxynaphthoyl moiety in DNA binding. Interestingly, the kedarcidin chromophore appears structurally related to neocarzinostatin yet recognizes specific DNA sequences in a manner similar to calicheamicin gamma 1I, an enediyne with a significantly different structure. Moreover, kedarcidin and calicheamicin share a DNA preferred site, the TCCTN-mer. These observations indicate that the individual structural features of these agents are not solely responsible for their DNA selectivity. Rather, a complementarity between their overall tertiary structure and the local conformation of the DNA at the binding sites must play a significant role in the recognition process.
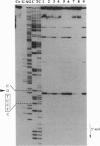
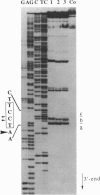
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boger D. L., Invergo B. J., Coleman R. S., Zarrinmayeh H., Kitos P. A., Thompson S. C., Leong T., McLaughlin L. W. A demonstration of the intrinsic importance of stabilizing hydrophobic binding and non-covalent van der Waals contacts dominant in the non-covalent CC-1065/B-DNA binding. Chem Biol Interact. 1990;73(1):29–52. doi: 10.1016/0009-2797(90)90107-x. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P., Heinemann U., Nurislamov A. A., Zielenkiewicz P., Dickerson R. E., Saenger W. Molecular dynamics simulation of the hydration shell of a B-DNA decamer reveals two main types of minor-groove hydration depending on groove width. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):593–597. doi: 10.1073/pnas.88.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedon P. C., Goldberg I. H. Influence of thiol structure on neocarzinostatin activation and expression of DNA damage. Biochemistry. 1992 Feb 25;31(7):1909–1917. doi: 10.1021/bi00122a003. [DOI] [PubMed] [Google Scholar]
- Drak J., Iwasawa N., Danishefsky S., Crothers D. M. The carbohydrate domain of calicheamicin gamma I1 determines its sequence specificity for DNA cleavage. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7464–7468. doi: 10.1073/pnas.88.17.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Giloni L., Takeshita M., Johnson F., Iden C., Grollman A. P. Bleomycin-induced strand-scission of DNA. Mechanism of deoxyribose cleavage. J Biol Chem. 1981 Aug 25;256(16):8608–8615. [PubMed] [Google Scholar]
- Goldberg I. H. Free radical mechanisms in neocarzinostatin-induced DNA damage. Free Radic Biol Med. 1987;3(1):41–54. doi: 10.1016/0891-5849(87)90038-4. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Alings C. Crystallographic study of one turn of G/C-rich B-DNA. J Mol Biol. 1989 Nov 20;210(2):369–381. doi: 10.1016/0022-2836(89)90337-9. [DOI] [PubMed] [Google Scholar]
- Hensens O. D., Dewey R. S., Liesch J. M., Napier M. A., Reamer R. A., Smith J. L., Albers-Schönberg G., Goldberg I. H. Neocarzinostatin chromophore: presence of a highly strained ether ring and its reaction with mercaptan and sodium borohydride. Biochem Biophys Res Commun. 1983 Jun 15;113(2):538–547. doi: 10.1016/0006-291x(83)91759-x. [DOI] [PubMed] [Google Scholar]
- Hensens O. D., Goldberg I. H. Mechanism of activation of the antitumor antibiotic neocarzinostatin by mercaptan and sodium borohydride. J Antibiot (Tokyo) 1989 May;42(5):761–768. doi: 10.7164/antibiotics.42.761. [DOI] [PubMed] [Google Scholar]
- Hofstead S. J., Matson J. A., Malacko A. R., Marquardt H. Kedarcidin, a new chromoprotein antitumor antibiotic. II. Isolation, purification and physico-chemical properties. J Antibiot (Tokyo) 1992 Aug;45(8):1250–1254. doi: 10.7164/antibiotics.45.1250. [DOI] [PubMed] [Google Scholar]
- Hurley L. H. DNA and associated targets for drug design. J Med Chem. 1989 Sep;32(9):2027–2033. doi: 10.1021/jm00129a001. [DOI] [PubMed] [Google Scholar]
- Kappen L. S., Ellenberger T. E., Goldberg I. H. Mechanism and base specificity of DNA Breakage in intact cells by neocarzinostatin. Biochemistry. 1987 Jan 27;26(2):384–390. doi: 10.1021/bi00376a008. [DOI] [PubMed] [Google Scholar]
- Kappen L. S., Goldberg I. H. Deoxyribonucleic acid damage by neocarzinostatin chromophore: strand breaks generated by selective oxidation of C-5' of deoxyribose. Biochemistry. 1983 Oct 11;22(21):4872–4878. doi: 10.1021/bi00290a002. [DOI] [PubMed] [Google Scholar]
- Konishi M., Ohkuma H., Matsumoto K., Tsuno T., Kamei H., Miyaki T., Oki T., Kawaguchi H., VanDuyne G. D., Clardy J. Dynemicin A, a novel antibiotic with the anthraquinone and 1,5-diyn-3-ene subunit. J Antibiot (Tokyo) 1989 Sep;42(9):1449–1452. doi: 10.7164/antibiotics.42.1449. [DOI] [PubMed] [Google Scholar]
- Kozarich J. W., Worth L., Jr, Frank B. L., Christner D. F., Vanderwall D. E., Stubbe J. Sequence-specific isotope effects on the cleavage of DNA by bleomycin. Science. 1989 Sep 22;245(4924):1396–1399. doi: 10.1126/science.2476851. [DOI] [PubMed] [Google Scholar]
- Lam K. S., Hesler G. A., Gustavson D. R., Crosswell A. R., Veitch J. M., Forenza S., Tomita K. Kedarcidin, a new chromoprotein antitumor antibiotic. I. Taxonomy of producing organism, fermentation and biological activity. J Antibiot (Tokyo) 1991 May;44(5):472–478. doi: 10.7164/antibiotics.44.472. [DOI] [PubMed] [Google Scholar]
- Long B. H., Golik J., Forenza S., Ward B., Rehfuss R., Dabrowiak J. C., Catino J. J., Musial S. T., Brookshire K. W., Doyle T. W. Esperamicins, a class of potent antitumor antibiotics: mechanism of action. Proc Natl Acad Sci U S A. 1989 Jan;86(1):2–6. doi: 10.1073/pnas.86.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marky L. A., Kupke D. W. Probing the hydration of the minor groove of A.T synthetic DNA polymers by volume and heat changes. Biochemistry. 1989 Dec 26;28(26):9982–9988. doi: 10.1021/bi00452a016. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Sugiura Y., Shiraki T., Konishi M., Oki T. DNA intercalation and cleavage of an antitumor antibiotic dynemicin that contains anthracycline and enediyne cores. Proc Natl Acad Sci U S A. 1990 May;87(10):3831–3835. doi: 10.1073/pnas.87.10.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura Y., Uesawa Y., Takahashi Y., Kuwahara J., Golik J., Doyle T. W. Nucleotide-specific cleavage and minor-groove interaction of DNA with esperamicin antitumor antibiotics. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7672–7676. doi: 10.1073/pnas.86.20.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S., Landovitz R., Ding W. D., Ellestad G. A., Kahne D. Cleavage behavior of calicheamicin gamma 1 and calicheamicin T. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4608–4612. doi: 10.1073/pnas.89.10.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zein N., Poncin M., Nilakantan R., Ellestad G. A. Calicheamicin gamma 1I and DNA: molecular recognition process responsible for site-specificity. Science. 1989 May 12;244(4905):697–699. doi: 10.1126/science.2717946. [DOI] [PubMed] [Google Scholar]
- Zein N., Sinha A. M., McGahren W. J., Ellestad G. A. Calicheamicin gamma 1I: an antitumor antibiotic that cleaves double-stranded DNA site specifically. Science. 1988 May 27;240(4856):1198–1201. doi: 10.1126/science.3240341. [DOI] [PubMed] [Google Scholar]