Fig 2. Multiple sequence alignment of MCF1-SHE domain proteins.
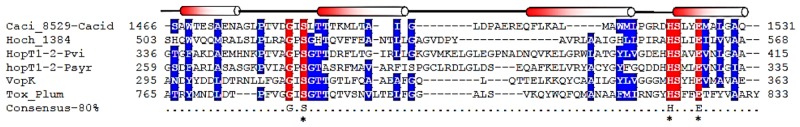
Alignment of VopK along with other polymorphic toxins and effector proteins as carried out using T-coffee [36]. The conserved and predicted SHE motif is marked with star (*). Identical and similar regions are shown highlighted as red and blue, respectively. Helices in the secondary structure of VopK as predicted by PSI-PRED [37, 38] are shown as cylinders (red and white gradient) along with coils as lines (black). Secondary elements of VopK are overlapping with those of other proteins as shown by Arvind and colleagues [9]. Accession numbers are as follows: ACU77352,Caci_8529 [Catenulispora acidiphila DSM 44928]; ACY13938, Hoch_1384 [Haliangium ochraceum DSM 14365]; EEA94476, HopT1-2 [Pseudovibrio sp. JE062]; EEB62143, hopT1-2 [Pseudomonas syringae pv. tomato T1]; EDN16474, VopK A33_1699 [Vibrio cholerae AM-19226]; AAM88787, Toxin protein [Photorhabdus luminescens].
