Fig. 1.
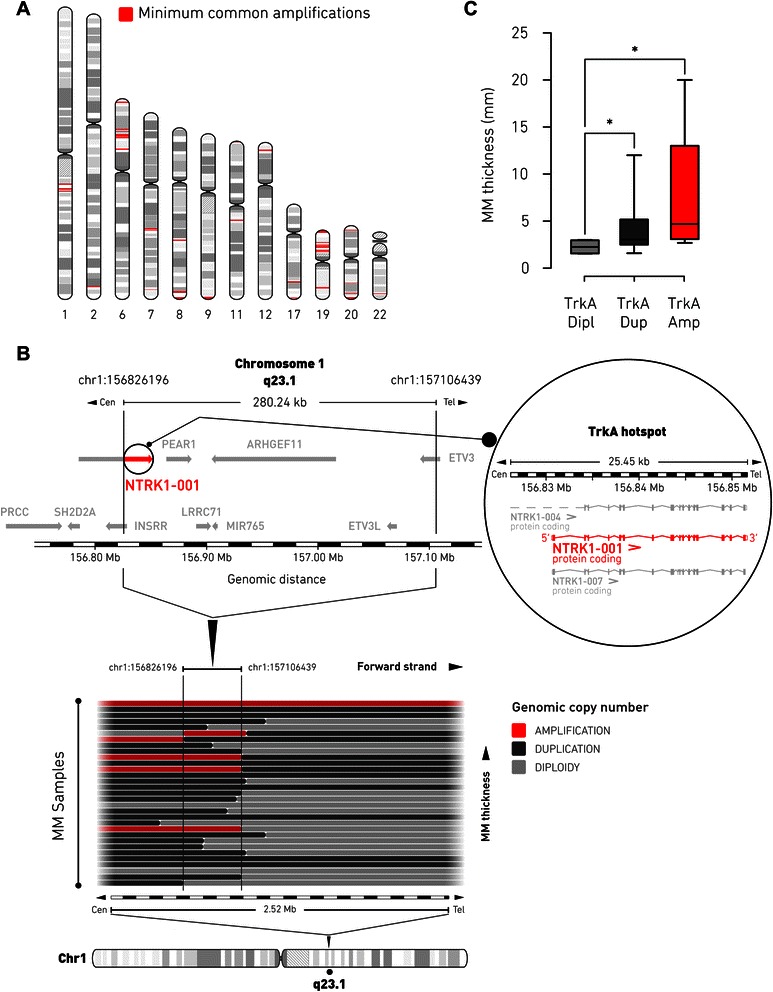
Identification of TrkA-1q23.1 genomic amplification in MM patients. a, hotspots of 40 minimum common amplifications (red) in primary MM genome, detected by aCGH across 31 patient samples, are plotted along their corresponding chromosome position and proportionally to the respective amplicon size. Detailed genomic information of hotspots is provided in Additional file 1: Table S2. b, schematic segmental gain profile within the 1q23.1 region (spanning ~2.5 Mb), as defined by aCGH, is represented with horizontal bars, each denoting the copy number status of an individual MM patient. MM samples with increasing primary tumor thickness are at the top (for details see Additional file 1: Table S4). Genomic amplifications are depicted in red. The black boundaries delineate the extent of the minimal common amplification (genomic coordinates chr1:156826196 to chr1:157106439). The graphical layout of the genes localized in the minimal amplification is based on the Ensembl release 75.37 of the human genetic map. The region of minimal common amplification extends over ~280 kb and retains the functional transcript of the NTRK1 gene [GenBank: Y09028], NTRK1-001 (red and inset), which codes for TrkA protein. For each panel, the corresponding scale of genomic positions (in Mb) is indicated. c, box and whiskers graph showing the association of TrkA-1q23.1 minimal amplification and tumor thickness in primary MM samples analyzed by aCGH (n = 31; Mann-Whytney U test: *, p < 0.05). Dipl, diploid copy number; Dup, duplication; Amp, amplification
