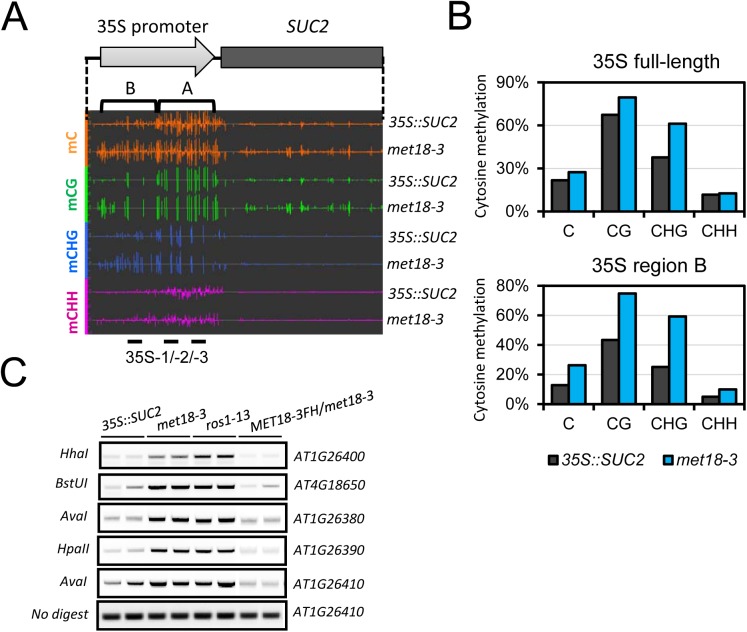
Fig 2. Epigenetic modification changes of the 35S::SUC2 transgene promoter in met18-3 mutant.
A. Diagram of the 35S::SUC2 transgene and an IGB snapshot of DNA methylation levels in the promoter region in different cytosine contexts. The positions of primer pairs used in ChIP-qPCR are as labeled. B. DNA methylation levels in the promoter region of 35S::SUC2. DNA methylation levels were quantified based on whole-genome bisulfite sequencing data. C. DNA methylation-sensitive PCR (Chop-PCR) results showing increased DNA methylation in met18-3 and ros1-13 mutants. Complementation with MET18 genomic sequence rescued increased DNA methylation. Non-digested DNA was used as the control PCR template.