Figure 1.
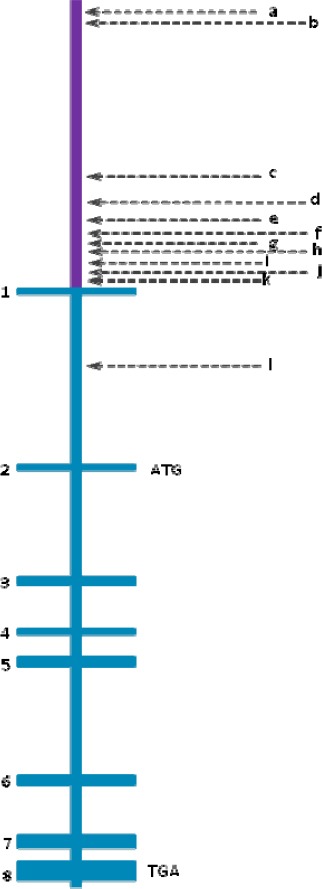
Pigment epithelium-derived factor (PEDF) gene structure containing its promoter region and its eight exons and seven introns
-
▪Promoter is represented in purplemmmm
-
▪a, b and c represent three different putative response elements to p63 y p73, which are located at -6362, -5996, and -1366
-
▪d, f and h are binding sites for transcription factors, located at ALU sequences (-682, -480, and -150). The most striking and unique feature of this region is the dense cluster of Alu elements which comprises 70% of the most proximal 1 kb upstream region. A sequence identical to Alu subclass (consensus sequence GGTCA(n)3 TGGTC(n)9 TGACC), which can function as an estrogen receptor-dependent transcriptional enhancer, is present in the PEDF upstream Alu repeat and a sequence differing by only one nucleotide is present in the proximal Alu repeat, i.e., within 200 bps of the translational start site141.
-
▪e is a sequence that is recognized by the C/EBP (CAAT-enhancer binding protein) family of transcription factors
-
▪g represents a sequence contained within TREp (palindromic thyroid hormone-responsive element). It is similar to the developmentally-regulated RAR (retinoic acid receptor) (-204).
-
▪i is the sequence for PEA3 (polyomavirus enhancer activator-3). It is present in tandem at -122,-129 and again at -141
-
▪j is a binding element for transcription factors such as the Oct (octamer-binding factor) family (-113)
-
▪k represents binding sites for CAAT-enhancer-binding proteins, C/EBPs or CHOP (-40), HNF4 (hepatocyte nuclear factor 4) (-60) and USF (upstream stimulatory factor)
-
▪l represents the MITF binding site (intron 1)
- ▪
