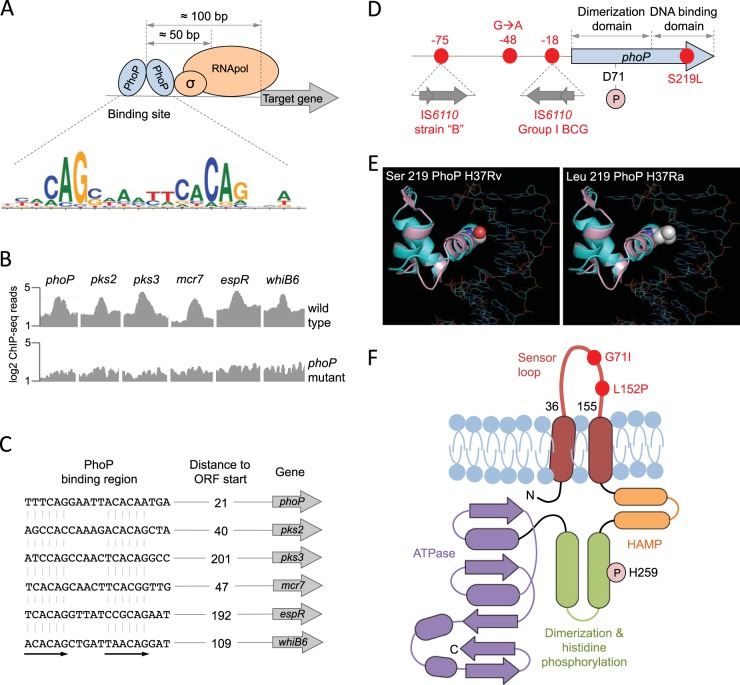
FIG 2 .
Molecular characteristics of the PhoPR two-component system. (A) Positioning of the PhoP transcription factor relative to the RNA polymerase and its target genes measured as the most probable values from ChIP-seq data. The PhoP consensus motif [TCACAG(N5)TCACAG] is also indicated. (B) ChIP-seq reads from representative promoters of PhoP-regulated genes. Note the significant increase in ChIP-seq peaks in the wild-type strain relative to those in its phoP mutant, indicative of a specific interaction of PhoP with these regions. (C) PhoP binding motifs elucidated from ChIP-seq data from the analyses whose results are shown in panel B. Distances to the start codons of target genes are also indicated. ORF, open reading frame. (D) Genomic locations of relevant phoP polymorphisms discussed throughout the text. Insertions of IS6110 in the promoter region and their positions relative to the phoP start codon are shown. Amino acid positions of the Asp71 residue, which is involved in the phosphotransfer reaction, and the Ser219Leu substitution in H37Ra are also indicated. (E) Ribbon models of the DNA binding domain of PhoP superimposed over the structure of a PhoB-DNA complex. Solid spheres show the wild-type serine 219 in H37Rv (left) or the leucine residue that appears in H37Ra (right) (adapted from reference 40). The mutant leucine residue is expected to interfere with DNA binding and/or recognition. (F) Secondary structure of the PhoR sensor kinase indicating its membrane topology. Each domain has been colored individually, and α-helices (ovals) and β-strands (arrows) are indicated. Note the presence of PhoR polymorphisms in the sensor loop, which is located in the periplasmic space. The position of the histidine involved in the phosphotransfer reaction is also indicated.