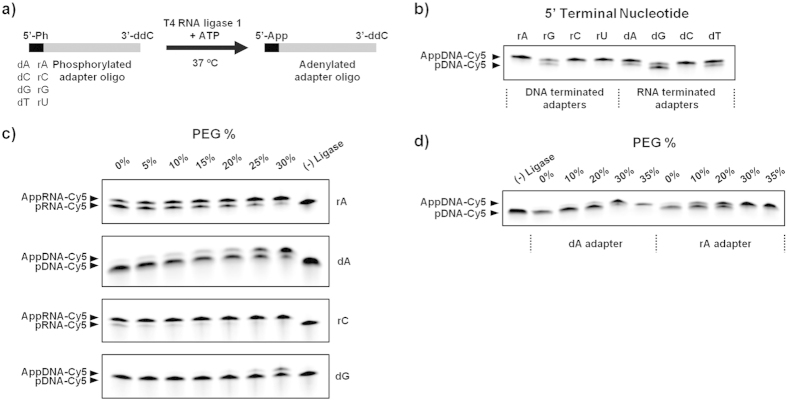
Figure 1.
(a) Schematic illustration of the high efficiency, purification- and template-free, adapter oligonucleotide adenylation method using T4 RNA ligase 1. The 3′ end of the adapter oligo was blocked by –ddC modification to prevent circularization and concatemerization. The 5′ base (shown in black) was swapped between dA, dC, dG, dT, rA, rC, rG, and rU to test bias. (b) The adapter adenylation efficiency was investigated as a function of 5′ terminal nucleotide. The reaction conditions were modified to exaggerate differences in efficiency (10 μL volume, 100 units ligase per nanomole adapter, 0.1 nanomole adapter, 30% PEG, 1 hour incubation). The rC and dG adapters are the most and least efficiently adenylated, respectively. (c) The adapter adenylation efficiency was then measured as a function of PEG % for a few representative adapters. In all cases, efficiency monotonically increased with PEG %. (d) Comparison of adenylation efficiency of as a function of PEG % under standard reaction conditions using the rA and dA adapters. Both the dA and rA adapters are efficiently adenylated at 35% PEG.