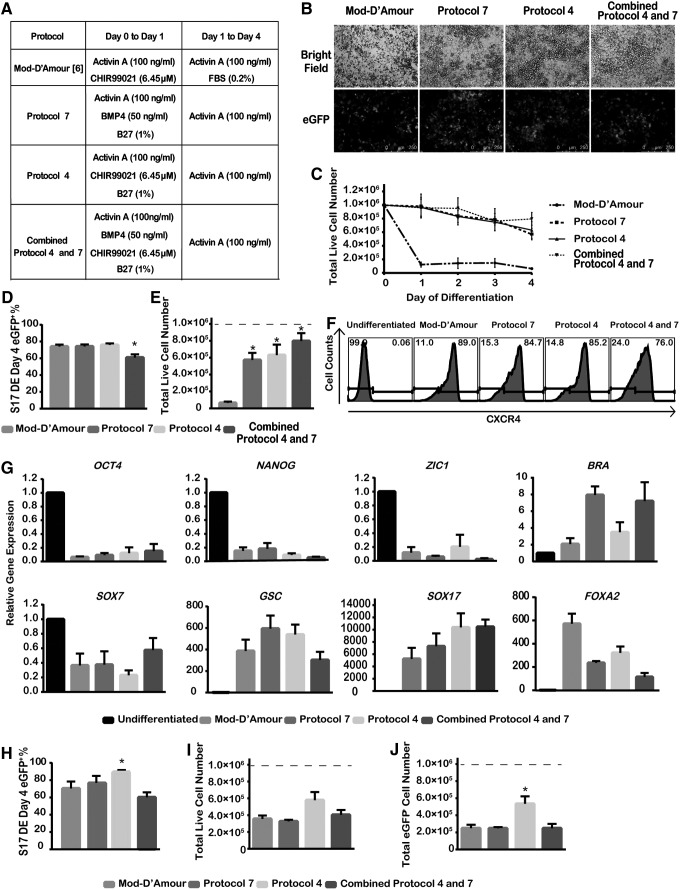
FIG. 2.
Three modified protocols for DE differentiation. (A) Summary of control and three modified DE differentiation protocols. Supplemented R medium (comprising 1% nonessential amino acids, 2 mM l-glutamine, 50 units/mL of penicillin, and 50 μg/mL of streptomycin in RPMI-1640 medium) was added to induce differentiation on a daily basis. (B) Bright field and immunofluorescence of eGFP+ cells on day 4 of S17d5 cell DE differentiation. Scale bar, 250 μm. (C) Daily total live cell number changes during S17d5 cell DE differentiation. (D) The percentage of eGFP+ cells after 4-day DE differentiation of S17d5 cells. (E) Total live cell numbers at the end of 4-day DE differentiation of S17d5 cells. (F) Representative flow cytometric histograms of CXCR4 induction after 4-day DE differentiation of S17d5 cells. (G) Gene expression of lineage-specific markers on day 4 of DE differentiation. OCT4 and NANOG, pluripotency markers; ZIC1, ectoderm marker; Brachyury (BRA), mesoderm marker; SOX7, visceral endoderm marker; GSC, mesendoderm marker; SOX17 and FOXA2, endoderm markers. (H–J) Three modified protocols for DE differentiation in feeder-free condition. Independent runs were normalized to 1.0×106 starting undifferentiated cells (dotted line). *P<0.05 compared with the Mod-D'Amour protocol by Student's unpaired t-test. Data are presented as mean±SEM, n=3–5.