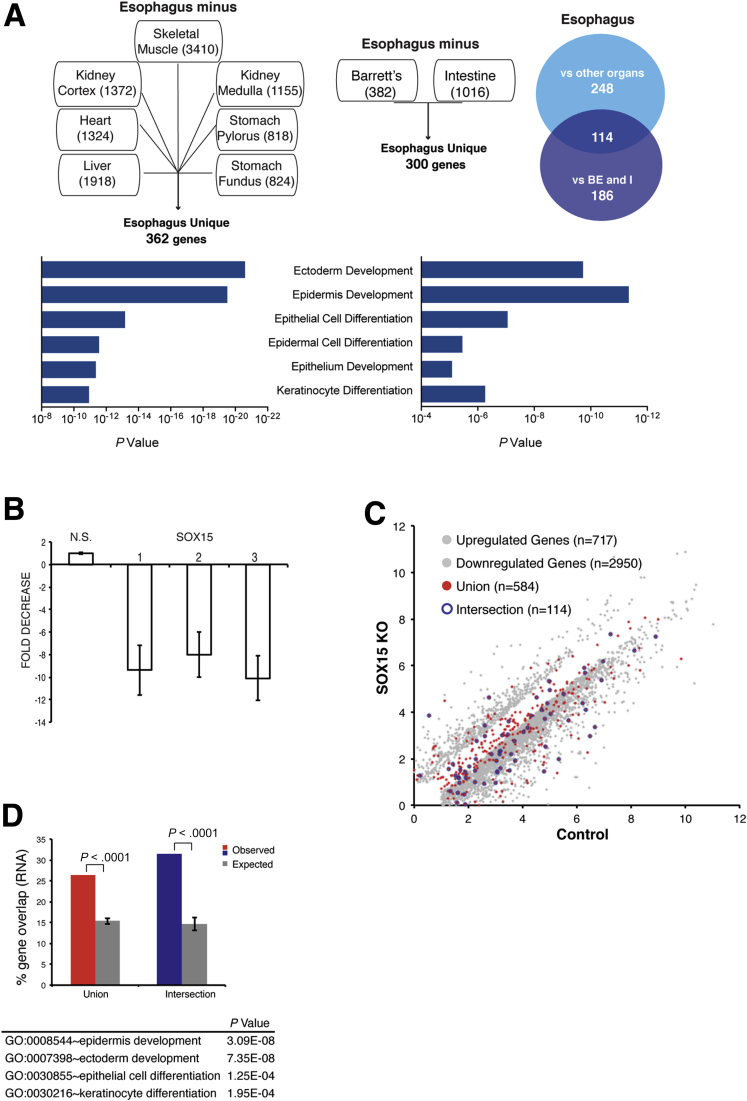
Figure 3.
Impact of SOX15 depletion on esophageal gene expression. (A) Delineation of the human esophageal transcriptome. The mRNAs expressed in human esophageal necropsy specimens (left) were compared against transcripts from 7 other postmortem organs,21 and mRNAs present in fresh esophageal biopsy specimens (right) were compared against transcripts from fresh Barrett’s esophagus (BE) and intestinal biopsies.8 Numbers in each box represent squamous esophagus-specific genes relative to that tissue. We identified 362 and 300 esophagus-specific genes, respectively, with a significant 114-gene overlap (P < .0001, chi-square test). The top Gene Ontology (GO) terms in each case are highly related to stratified epithelia. (B) SOX15 mRNA depletion in 3 representative experiments in which CPA cells were infected with lentiviruses carrying SOX15-specific or a nonspecific (NS) 21-bp shRNAs. Knockdown efficiency, assessed by quantitative reverse-transcription polymerase chain reaction 72 hours after infection, was >8- to 10-fold in every experiment. (C) Results of duplicate RNA-seq analysis of genes differentially expressed in CP-A cells treated with SOX15-specific (y-axis) or control, nonspecific (x-axis) shRNAs. Grey dots mark differential expression (log2 >1.5-fold, q < .05); genes present in the union (548 genes) or intersection (114) sets of esophagus-specific genes are represented by red and blue dots, respectively. (D) Fraction of esophagus-specific transcripts reduced upon SOX15 depletion (red, 548 union-set genes; blue, 114 intersection-set genes) compared with five random sets of equal numbers of genes expressed in CP-A cells (grey bars). The table lists GO terms enriched among SOX15-dependent genes.