Fig. 8.
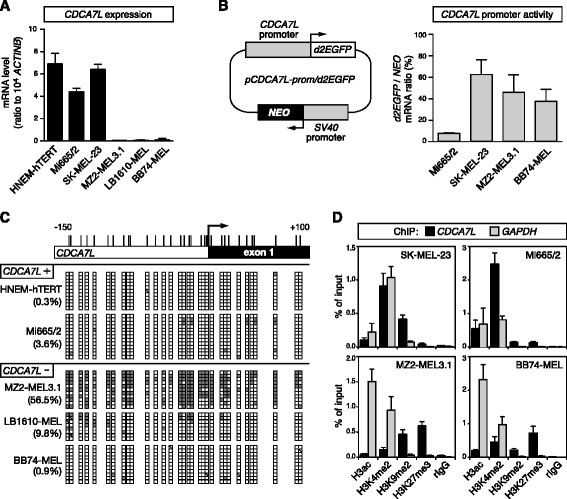
Epigenetic mechanisms associated with CDCA7L repression in melanoma cells. a CDCA7L mRNA expression levels were assessed in immortalized normal melanocytes (HNEM-hTERT) and in five melanoma cell lines (Mi665/2 and SK-MEL-23, MZ2-MEL3.1, LB1610-MEL, and BB74-MEL). Values represent mean (± sem) of at least two independent RT-qPCR experiments, each in duplicate. b Schematic representation of the pCDCA7L-prom/d2EGFP plasmid (left panel). Four melanoma cell lines were stably transfected with the pCDCA7L-prom/d2EGFP plasmid, and d2EGFP mRNA expression levels were evaluated (right panel). Results were normalized with NEO gene expression levels. Data represent mean (± sem) of three independent RT-qPCR analyses, each in duplicate. c Bisulfite sequencing analyses of the CDCA7L 5'-region were performed in HNEM-hTERT cells and in four melanoma cell lines displaying CDCA7L expression or repression (CDCA7L+ or −, respectively). Methylated and unmethylated CpGs are represented by filled and empty boxes, respectively. Overall CpG methylation percentages (%) are indicated. d Quantitative ChIP analyses were applied to four melanoma cell lines displaying either CDCA7L expression (SK-MEL-23, Mi665/2) or repression (MZ2-MEL3.1, BB74-MEL). Enrichment of indicated histone modifications were evaluated within the CDCA7L 5′-region (GAPDH served as a control, ubiquitously active, promoter). Data derive from at least two independent ChIP experiments, with two duplicate qPCR measures in each case
