Fig. 2.
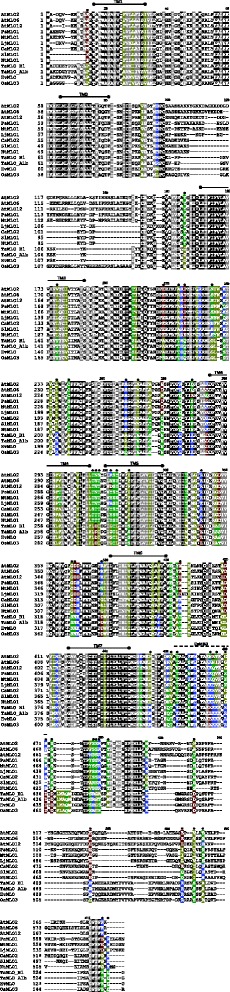
Multiple alignment of MLO powdery mildew susceptibility proteins. The dataset is composed of all the monocot (barley HvMLO, rice OsMLO3, wheat TaMLO_B1 and TaMLO_A1b), and dicot (Arabidopsis AtMLO2, AtMLO6 and AtMLO12, tomato SlMLO1, pepper CaMLO2, tobacco NtMLO1, pea PsMLO1, lotus LjMLO1 and barrel clover MtMLO1) MLO homologs shown to act as powdery mildew susceptibility factors. The positions of the seven MLO transmembrane domains (TM1-TM7) and the calmodulin binding domain (CaMBD) are identical to the ones reported by Feechan et al. [2], Functional Plant Biology, 35: 1255–1266. Black color indicates alignment positions in which invariable residues are present. Grey color indicates alignment positions which do not contain class-specific residues and are conserved with respect to biochemical properties. Other colors indicate alignment positions in which there are class-specific residues in monocots, dicots, or both: yellow indicates hydrophobic residues (G, A, V, L, I, F, W, M, P); blue indicates polar basic residues (K,R,H); red indicates polar acidic residues (D, E); green indicates polar uncharged residues (S, T, C, Y, N, Q). Black dots highlight 44 alignment positions in which class-specific residues are substituted in the other class by residue(s) having different biochemical properties
