Fig. 3.
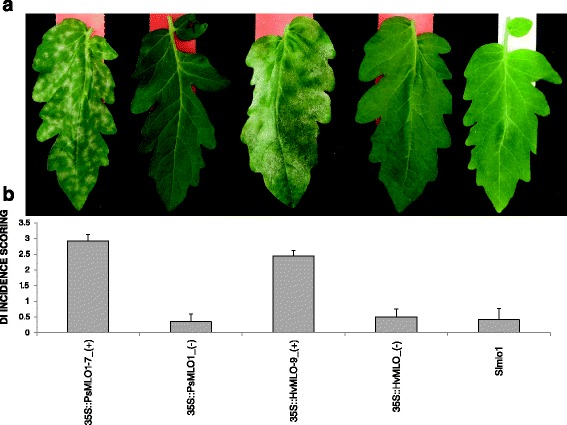
Transgenic overexpression of pea PsMLO1 and barley HvMLO in the tomato mutant line Slmlo1. Panel a shows the phenotypes of two selected individuals of the T2 family 35S::PsMLO1-7, segregating for the presence (first from the left) or the absence (second from the left) of the transgene, two selected individuals of the T2 family 35S::HvMLO-9, segregating for the presence (third from the left) or the absence (second from the right) of the transgene, and one individual of the Slmlo1 line (first from the right), in response to the tomato powdery mildew fungus Oidium neolycopersici. Panel b from left to right shows average disease index (DI) values relative to transgenic plants (+) of the 35S::PsMLO1-7 T2 family, non-transgenic plants (−) of three T2 families segregating for the 35S::PsMLO1 construct, transgenic plants of the 35S::HvMLO-9 T2 family, non-transgenic plants of three T2 families segregating for the 35S::HvMLO construct and the Slmlo1 line. Standard deviation bars refer to six 35S::PsMLO1_(+) individuals, nine 35S::HvMLO_(+) individuals, 7 PsMLO1_(−) individuals, 7 HvMLO_(−) individuals and 10 Slmlo1 individuals
