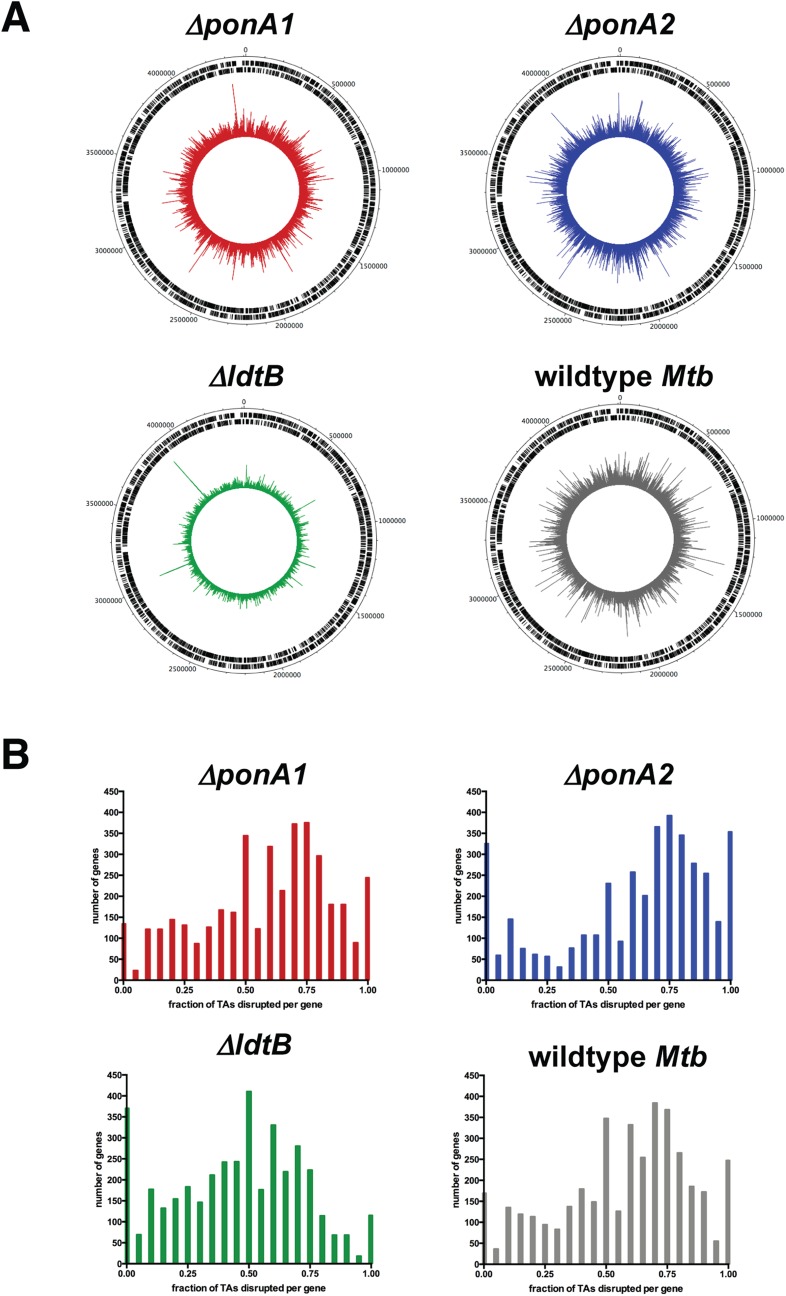
Fig. S2.
The transposon libraries are highly saturated. (A) The chromosomal distribution of sequence reads from the libraries was visualized with DNAPlotter. (B) The fraction of possible insertion sites (TA dinucleotides) that were actually disrupted by transposons was calculated per gene and plotted. The ΔponA1 library’s saturation is circa 70%. The ΔponA2 library is about 78% saturated. The ΔldtB library is circa 55% saturated, and wild-type Mtb is around 70% saturation.