Fig. S2.
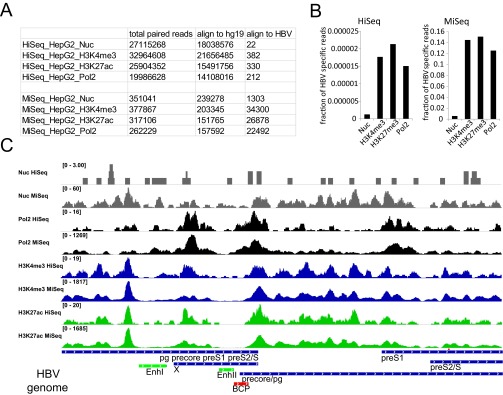
Controls for HBV target enrichment steps in HepG2-NTCP1 cccDNA ChIP-Seq assay. (A) Total number of paired reads from mononucleosomes (Nuc), H3K4me3, H3K27ac, and Polymerase 2 (Pol2) ChIP that align to human (hg19) and HBV genomes. Samples that were sequenced on a HiSeq2500 (HiSeq_X) instrument were not enriched for HBV DNA, whereas samples sequenced on a MiSeq instrument (MiSeq_X) were enriched for HBV DNA by target enrichment before sequencing. The same data are presented in B for HiSeq (Left) and MiSeq (Right) as the fraction of all mapped paired reads that align to the HBV genome. For sufficient sequencing coverage 1,455 paired mapped reads would be required (SI Materials and Methods). (C) Distribution of nucleosome, RNA Polymerase 2, H3K4me3, and H3K27ac read density along the HBV genome. The difference in the total number of reads is indicated in the upper left corner of each sequencing track.
