FIGURE 1.
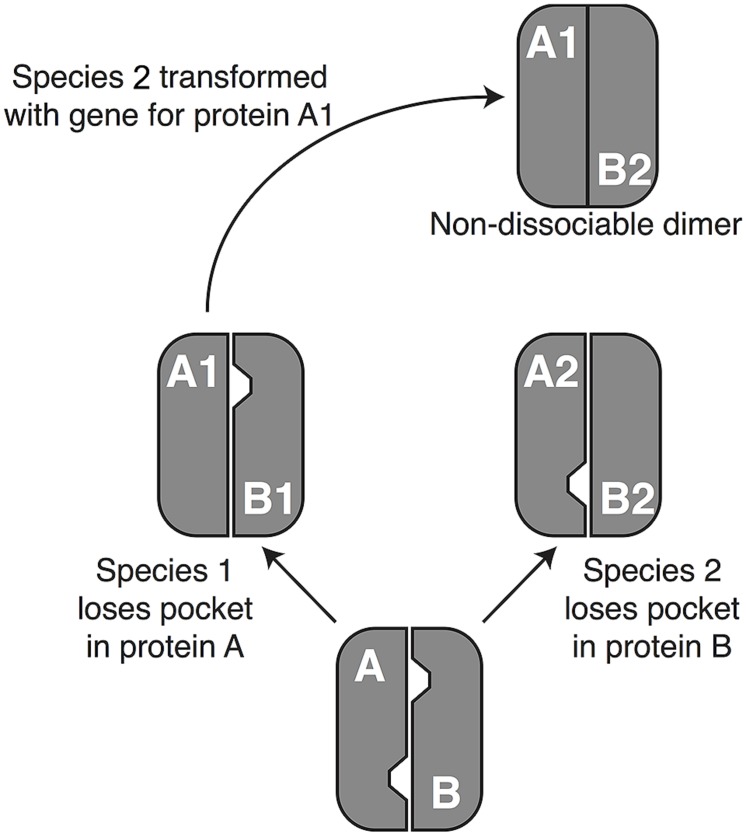
A possible mechanism of development system drift (DSD). DSD occurs when two lineages remain phenotypically unchanged but undergo genetic divergence (True and Haag, 2001). The phenomenon depends primarily on evolutionary changes that influence the way that proteins interact with each other and with DNA sequences (Haag, 2007; Landry et al., 2007), although changes in miRNA’s and their targets (Mallory et al., 2004) have a potential role as well. To understand how such phenomena can lead to a dominant phenotype in a transgenomic screen, consider a hypothetical example in which two proteins, A and B, dimerize but need to dissociate during normal development. In a hypothetical ancestor, pockets in both proteins destabilize the dimer enough to permit dissociation. In the lineage leading to species 1, protein A (A1) loses its pocket, but dissociation is still achieved thanks to the pocket in protein B1. Conversely, the pocket in protein B2 has been lost on the lineage leading to species 2. Moving protein A1 in species 2 (or B2 into species 1) will cause formation of a non-dissociable dimer, resulting in a dominant disruption of normal development. When development is disrupted sufficiently to cause inviability or sterility, DSD can enforce reproductive isolation between lineages, because hybrids do not survive to reproduce. In that case the pattern conforms to the Dobzhansky–Muller model of speciation [e.g., (Bomblies et al., 2007; Landry et al., 2007)], showing that transgenomics offers a novel way to identify genetic interactions that could contribute to speciation.