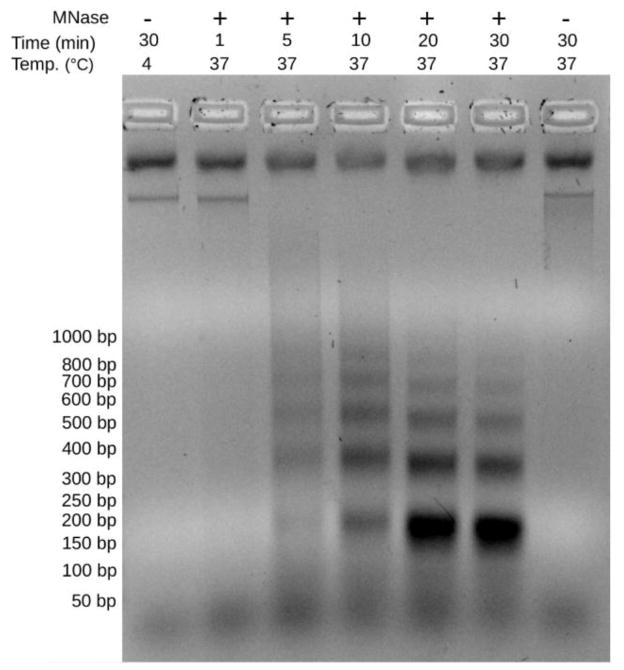
Fig. 2.
Optimization of micrococcal nuclease (MNase) digestion. Many different conditions were tested for the three Zymoseptoria species, the best being 20 minutes at 37°C. Shown here is MNase digestion of Z. ardabiliae chromatin with either 1000 units of MNase (“+”; New England Biolabs) or no enzyme (“−”) at 4°C and 37°C. The digestion time was optimized to release mostly mononucleosomes (~150 bp). Digestion profiles for Z. tritici and Z. pseudotritici revealed similar optimal conditions (data not shown). Optimal digestion can vary for different species and strains and should be done and re-checked for each different set of strains.