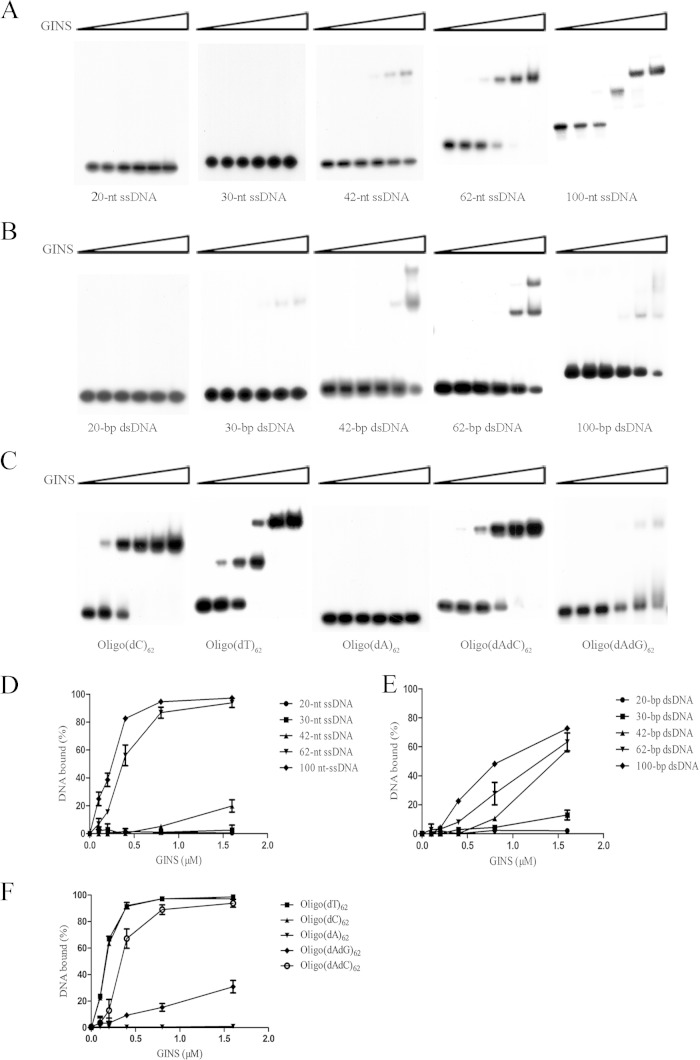
FIG 1.
Binding of SsoGINS to ssDNA and dsDNA fragments. (A, B, D, and E) Binding of SsoGINS to ssDNA or dsDNA of various lengths. SsoGINS was mixed with 32P-labeled 20-, 30-, 42-, 62-, and 100-nt ssDNA fragments (S1, S3, S7, S17, and OligoA) (see Table S1 in the supplemental material) or with 20-, 30-, 42-, 62-, and 100-bp dsDNA fragments (D1, D8, D9, D7, and D10) (see Table S1). The protein-DNA complexes were subjected to polyacrylamide gel electrophoresis. The gel was exposed to X-ray film (A and B) and quantified by phosphorimaging (D and E). Concentrations of SsoGINS were 0, 0.1, 0.2, 0.4, 0.8, and 1.6 μM, indicated by the triangles above the lanes. (C and F) Sequence preference of SsoGINS in ssDNA binding. SsoGINS was mixed with a 32P-labeled 62-nt homopolymeric ssDNA or a 62-nt ssDNA with alternating A and G or A and C (see Table S1). The protein-DNA complexes were subjected to polyacrylamide gel electrophoresis. The gel was exposed to X-ray film (C) and quantified by phosphorimaging (F). Concentrations of SsoGINS used in the assays were 0, 0.1, 0.2, 0.4, 0.8, and 1.6 μM, in respective order, as indicated by the triangle above the lanes. Data shown in panels D to F represent an average of three independent measurements.