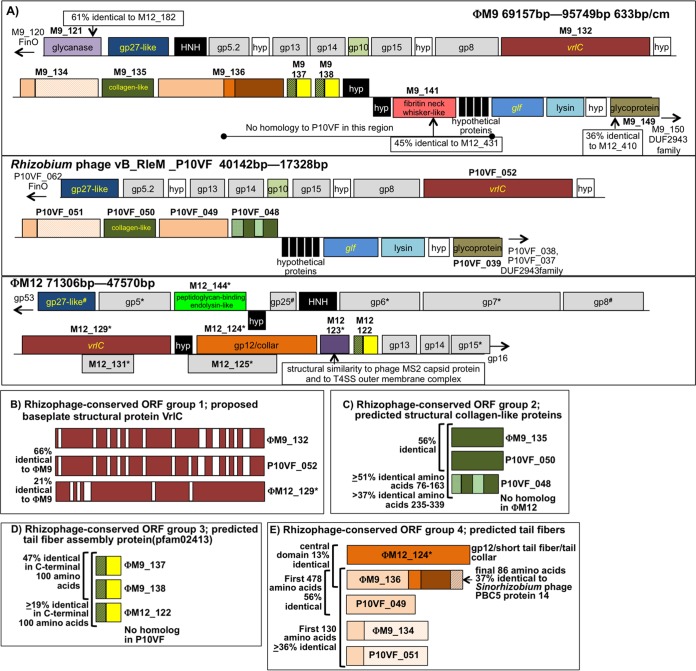
FIG 4.
(A) Detailed synteny in the neck/baseplate/tail region of the genomes of ΦM9, Rhizobium phage P10VF, and ΦM12. ORFs filled in black do not have homologs in either of the other two genomes. ORFs filled in white in ΦM9 and P10VF encode hypothetical proteins conserved at the same position in P10VF. The genomes are oriented in the same direction with respect to baseplate protein gp27. All three genomes have baseplate proteins gp5 and gp8 and neck proteins gp13, gp14, and gp15 shaded in gray. (Asterisks next to ΦM12 ORF names mean that these proteins were detected in the ΦM12 proteome [10].) There is nearly complete synteny between ΦM9 and P10VF, except for the absence from P10VF of the region from the middle of ΦM9_136 to the beginning of the glf gene (encodes UDP-galactopyranose mutase) and the absence from P10VF of the glycanase encoded by ΦM9_121. Four ΦM9 ORFs in the region shown lack homologs in P10VF but have homologs in ΦM12, i.e., ΦM9_121 (glycanase), ΦM9_141, and ΦM9_137, and ΦM9_138, which are partial duplicates of one another. (B) Rhizophage-conserved ORF group 1. Shown are the predicted VrlC proteins from the three phage genomes with regions of homology in red and gaps in white. VrlC is one of the more abundant proteins in the ΦM12 proteome (10). (C) Rhizophage-conserved ORF group 2, which is structurally similar to collagen (31) and consists of one ΦM9 ORF and two P10VF ORFs. Regions of homology are solid green. The two nearly adjacent P10VF ORFs may have arisen from a gene duplication (see panel A). (D) Rhizophage group 3, predicted tail fiber assembly proteins, is composed of two very similar adjacent ORFs in ΦM9, a homolog in ΦM12, and ORFs from five other phages of rhizobia (not pictured) (see the text and Table S4 in the supplemental material). (E) Rhizophage predicted tail fiber proteins, group 4. ΦM12_124 is the third most abundant protein in the ΦM12 proteome and is predicted to encode a T4 gp12-like/short-tail fiber/phage tail collar protein. ΦM9_136 shows 13% identity with ΦM12_124 in its central domain and has a C-terminal domain that is 37% identical to Sinorhizobium phage PBC5 protein 14 (GenBank accession no. NC_003324.1) and an N-terminal domain that is 56% identical to phage P10VF_049. ORFs ΦM9_136 and P10VF_049 are ≥36% identical in their first 130 amino acids to ΦM9_134 and P10VF_051, which are structurally similar to B. subtilis short-tailed phage neck appendage proteins (75, 76). These ORFs may have arisen from gene duplication events in the ΦM9 and P10VF genomes or in a common progenitor strain.