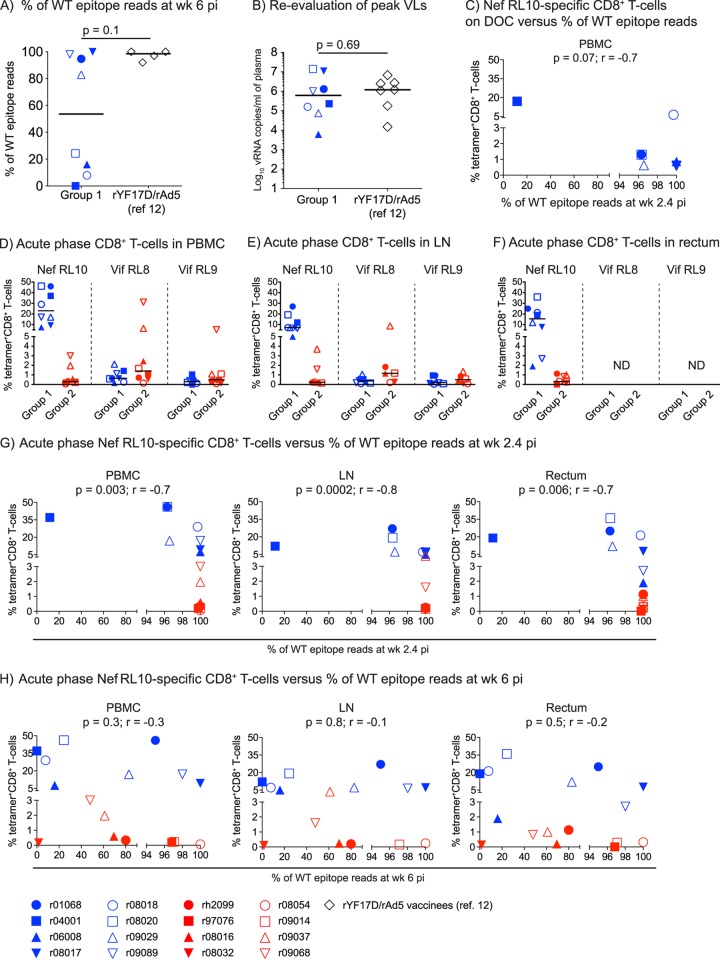
FIG 9.
Biological variables affecting the kinetics of CD8+ T-cell escape within Nef RL10. (A) Comparison of the frequencies of WT Nef RL10 sequence reads detected at week 6 p.i. between group 1 and the rYF17D/rAd5-immunized animals in our latest Mamu-B*08 trial (12). Data from only four rYF17D/rAd5 vaccinees were available for this comparison. (B) Reevaluation of peak viral loads in macaques in group 1 and the rYF17D/rAd5 vaccinees. To avoid interassay variability, we determined plasma virus concentrations on freshly thawed samples using the same quantitative-PCR conditions. We used the Mann-Whitney test to calculate P values for the analyses depicted in panels A and B. The lines represent medians. (C) Magnitudes of Nef RL10-specific CD8+ T cells in PBMC of the group 1 animals on the DOC compared to their corresponding frequencies of viral quasispecies encoding WT Nef RL10 (WT epitope reads) measured at week 2.4 p.i. (D to F) Magnitudes of CD8+ T cells specific for the Mamu-B*08-restricted Nef RL10, Vif RL8, and Vif RL9 epitopes present in PBMC (D), LN (E), and rectum (F) from animals in groups 1 and 2 at week 2.4 p.i. (G and H) Magnitudes of acute-phase Nef RL10-specific CD8+ T cells in PBMC, LN, and rectum from animals in groups 1 and 2 compared to their corresponding frequencies of WT epitope reads at week 2.4 (G) and 6 (H) p.i. We determined the correlation coefficients in panels C, G, and H by using the Spearman rank correlation test. Each symbol denotes one monkey. ND, not done.