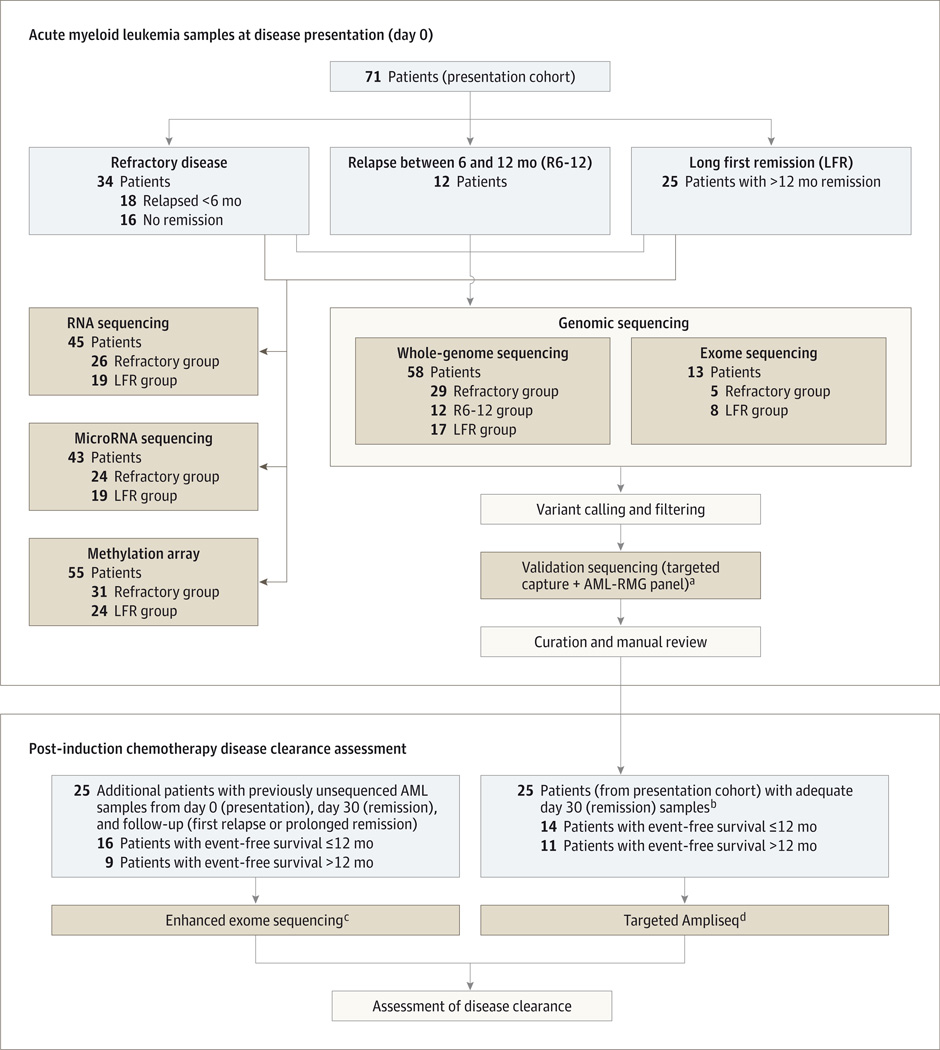
Figure 1.
Flowchart Outlining the Selection of Samples and Sequencing Approaches in the Study
aAML-RMG is a capture reagent consisting of all of the exons of the genes that are currently known to be recurrently mutated in acute myeloid leukemia, based on The Cancer Genome Atlas AML study.8
bThe only samples with sufficient day 30 DNA for sequencing and assessment of disease clearance (refractory group, 6 patients; R6-12 group, 8 patients; LFR group, 11 patients).
c Enhanced exome sequencing is exome capture-based sequencing supplemented with the AML-RMG panel of target genes, to improve coverage of critical regions of the exome.
d Targeted Ampliseq is a polymerase chain reaction–based digital sequencing approach that allows for accurate determination of the frequency of specific mutations in acute myeloid leukemia samples.