Abstract
Development of xylose-fermenting yeast strains that are tolerant to the inhibitors present in lignocellulosic hydrolysates is crucial to achieve efficient bioethanol production processes. In this study, the importance of the propagation strategy for obtaining robust cells was studied. Addition of hydrolysate during propagation of the cells adapted them to the inhibitors, resulting in more tolerant cells with shorter lag phases and higher specific growth rates in minimal medium containing acetic acid and vanillin than unadapted cells. Addition of hydrolysate during propagation also resulted in cells with better fermentation capabilities. Cells propagated without hydrolysate were unable to consume xylose in wheat straw hydrolysate fermentations, whereas 40.3% and 97.7% of the xylose was consumed when 12% and 23% (v/v) hydrolysate, respectively, was added during propagation. Quantitative polymerase chain reaction revealed changes in gene expression, depending on the concentration of hydrolysate added during propagation. This study highlights the importance of using an appropriate propagation strategy for the optimum performance of yeast in fermentation of lignocellulosic hydrolysates.
Introduction
The yeast Saccharomyces cerevisiae has been used for centuries by many cultures for baking and producing alcoholic beverages. Furthermore, during the past few decades, this yeast has been exploited to produce bioenergy – in particular, bioethanol.
The conversion of abundant lignocellulosic biomass into bioethanol is a sustainable alternative to the present industrial production of bioethanol, which uses starch and sucrose-derived feedstocks as raw materials. Fermentation of lignocellulose-derived materials is, however, very challenging for S. cerevisiae because apart from sugars, inhibitory compounds derived from cellulose, hemicellulose and lignin degradation during pretreatment of the biomass are also present in the resulting hydrolysate (Alvira et al., 2010). The inhibitory compounds prevent the growth of yeast and affect fermentation performance. Furthermore, wild-type S. cerevisiae is unable to ferment xylose, which can constitute up to 40% of the lignocellulosic material. Only enteric bacteria and some fungi and yeasts are able to ferment xylose but with low yield (Tomás-Pejó, 2011). Some ethanologenic bacteria like Escherichia coli have shown promising alternatives for industrial exploitation (Okuda et al., 2007). However, wild-type E. coli, for example, shows low ethanol yield because it converts sugar more efficiently to organic acids and several approaches have been performed with the aim of redirecting glycolytic fluxes to ethanol (Tao et al., 2001).
In order to upgrade the implementation of lignocellulosic bioethanol production to industrial scale, it is imperative to develop robust xylose-fermenting strains that work efficiently under the prevailing conditions. One commonly used strategy to enable wild-type S. cerevisiae to consume xylose is to introduce the XYL1 and XYL2 genes from Scheffersomyces stipitis. These genes encode xylose reductase (XR) and xylitol dehydrogenase (XDH) respectively. However, because of the different preferences of XR and XDH for co-factors, xylitol formation is one of the main drawbacks of using this strategy – leading to a lower yield of ethanol from xylose than from hexoses.
Because it has been shown that xylose fermentation capacity is more affected by inhibitors than glucose fermentation (Martín et al., 2007; Ask et al., 2013), the development of yeast strains that are tolerant to inhibitors is even more important when xylose-fermenting traits are introduced. During the last years, several studies involving metabolic and evolutionary engineering have been performed to obtain xylose-fermenting S. cerevisiae strains that work efficiently in lignocellulosic hydrolysates (Koppram et al., 2012; Kim et al., 2013; Gonçalves et al., 2014). In this context, it has been shown that evolutionary engineering (i.e. long-term adaptation) of xylose-fermenting strains to lignocellulosic hydrolysate not only leads to better results in terms of tolerance to inhibitors, but also to an increase in xylose fermentation capacity and ethanol yield compared with non-adapted cells (Martín et al., 2007; Tomás-Pejó et al., 2010).
Propagation of the seed culture is an important step in every fermentation process that demands efficient production of yeast cells with high fermentative efficiency. Exposing the cells to the inhibitory hydrolysate during the propagation step would allow short-term adaptation of the yeast cells to the inhibitors during pre-inoculum growth (Koppram et al., 2013). In this work, the effect of short-term adaptation during propagation on cell metabolism and gene expression is studied for the first time.
Molecular adaptation to different growth conditions during the propagation step is, however, poorly understood. In the last years, some studies have been carried out to analyse the complexity of the yeast biomass production process for wine strains (Gómez-Pastor et al., 2011), but little is known about how the propagation conditions influence the capacity of the cells to produce bioethanol from lignocellulosic materials.
In the present study, we assessed the importance of the propagation strategy for obtaining robust strains that efficiently co-ferment glucose and xylose in lignocellulosic hydrolysates. The engineered industrial strain S. cerevisiae KE6-12 harbouring the xylose genes (XR and XDH) from S. stipitis and overexpressing the endogenous xylulokinase was used as the fermenting microorganism. This strain was previously developed to grow well on lignocellulosic hydrolysates (Albers, E., Halpin, R. and Olsson, L. et al., unpublished) and has been successfully used in bioethanol production processes from lignocellulose (Moreno et al., 2013; Tomás-Pejó et al., 2014). However, in those cases, xylose consumption was not higher than 75% in 120–144 h, highlighting the importance of the propagation strategy to improve xylose consumption.
We studied the effect of the different propagation strategies on the specific growth rate in minimal medium with two inhibitory compounds (vanillin and acetic acid). Vanillin is a phenolic aldehyde compound that has been shown to be one of the most potent inhibitors as it inhibits fermentation at very low concentration (Klinke et al., 2004; Nguyen et al., 2014). On the other hand, acetic acid is a carboxylic acid commonly found in high concentration in lignocellulosic hydrolysates that contributes to cell arrest and reduction of ethanol productivities (Mira et al., 2010).
The performance of cells propagated under different conditions was compared during ethanol production processes from wheat straw hydrolysate. We also determined the influence that the addition of hydrolysate during propagation has on the expression of key genes known to be related to the stress response and tolerance to inhibitors.
Results and discussion
Propagation strategy
Depending on the propagation strategy, different concentrations of wheat straw hydrolysate were added during the cell propagation step. In the base case, no hydrolysate was used, but only a defined medium. The wheat straw hydrolysate was recovered after filtering the whole pretreated slurry. The latter had 20.4% (w/w) total solids and was obtained after pretreating the wheat straw at 190°C for 15 min in a steam explosion plant, by SEKAB in Örnsköldsvik, Sweden.
Wheat straw hydrolysate had the following composition: 14.5 g l−1 glucose, 32.6 g l−1 xylose, 3.5 g l−1 arabinose, 1.8 g l−1 mannose, 8.5 g l−1 acetic acid, 1.6 g l−1 formic acid, 1.3 g l−1 5-hydroxymethyl furfural (HMF), 7.7 g l−1 furfural and 0.05 g l−1 vanillin.
For the propagation, pre-inocula were grown at 30°C in 250 ml Erlenmeyer flasks shaken at 150 r.p.m. containing 50 ml of Delft medium as follows: 10 g l−1 glucose, 20 g l−1 xylose, 7.5 g l−1 (NH4)2SO4, 3.5 g l−1 KH2PO4, 0.75 g l−1 MgSO4·7H2O, 2 ml l−1 trace metal solution and 1 ml l−1 vitamin solution (Verduyn et al., 1990). Wheat straw hydrolysate, filtered and adjusted to pH 6, was added at the mid-exponential phase of pre-inoculum growth (OD600 nm ≈ 2.5) when the cells were starting to grow on xylose. According to the propagation strategy, wheat straw hydrolysate was added to reach a final concentration of 12% or 23% (v/v) in the pre-inoculum medium. The sugar content in 12% (v/v) hydrolysate was increased to have the same sugar content as present in 23% (v/v) hydrolysate. Furthermore, minimal medium only (designated 0% hydrolysate) with sugar content corresponding to that of 23% (v/v) was also used for comparative purposes.
After addition of hydrolysate, the pre-inoculum culture was grown for another 8 h. The specific growth rates for the propagation cultures were similar under the three conditions investigated.
Cell growth in the presence of acetic acid and vanillin
In order to check whether the propagation strategy had an effect on the tolerance to acetic acid and vanillin, the growth of cells propagated under different conditions was tested in 96 micro-well equipment Bioscreen C MBR (Growth Curves, Helsinki, Finland). The medium used was Delft minimal medium with 10 g l−1 glucose, 20 g l−1 xylose, 1 g l−1 vanillin, and 5 or 10 g l−1 acetic acid. The concentration of acetic acid was in the range of what would be predictable in steam-exploded wheat straw hydrolysates, but the concentration of vanillin (1 g l−1) was 10 times higher than what would be expected, in order to make the effect more evident (Tomás-Pejó et al., 2010).
It was clear that cells propagated with different hydrolysate concentrations (0, 12 or 23% (v/v)) had different growth curves in the presence of acetic acid and vanillin (Fig. 1).
Figure 1.
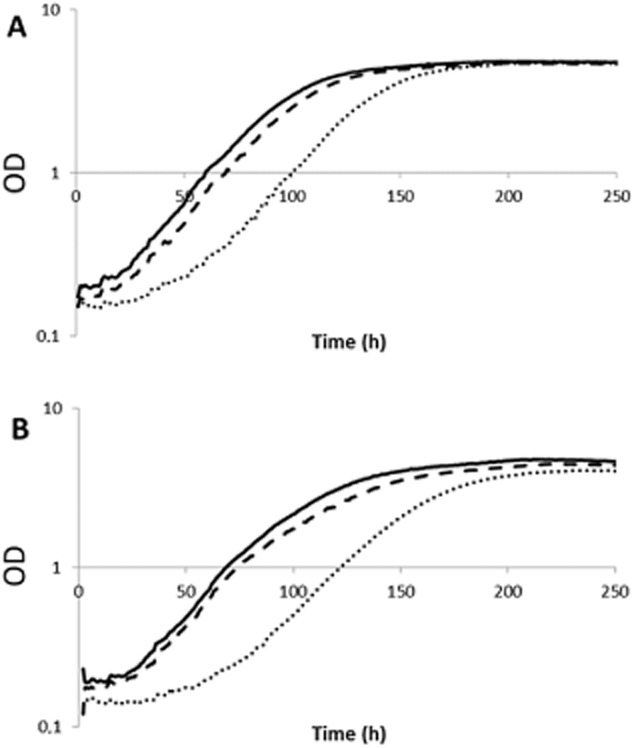
Growth in minimal medium with 10 g l−1 glucose and 20 g l−1 xylose; growth curves of cells propagated with ( ) 0% (v/v) hydrolysate, (
) 0% (v/v) hydrolysate, ( ) 12% (v/v) hydrolysate or (
) 12% (v/v) hydrolysate or ( ) 23% (v/v) hydrolysate in the presence of (A) 5 g l−1 acetic acid and 1 g l−1 vanillin or (B) 10 g l−1 acetic acid and 1 g l−1 vanillin. Ten growth curves were analysed for each condition for 3 days at 30°C in a working volume of 145 μl. Initial OD580 nm was 0.1. OD580 nm values were taken every 15 min using a wide-band filter with λ = 420–580 nm.
) 23% (v/v) hydrolysate in the presence of (A) 5 g l−1 acetic acid and 1 g l−1 vanillin or (B) 10 g l−1 acetic acid and 1 g l−1 vanillin. Ten growth curves were analysed for each condition for 3 days at 30°C in a working volume of 145 μl. Initial OD580 nm was 0.1. OD580 nm values were taken every 15 min using a wide-band filter with λ = 420–580 nm.
In minimal medium with 5 g l−1 acetic acid and 1 g l−1 vanillin (Fig. 1A), the specific growth rates were 0.02 h−1, 0.08 h−1 and 0.10 h−1 with cells propagated at 0%, 12% and 23% (v/v) hydrolysate respectively. On the other hand, the specific growth rates in the presence of 10 g l−1 acetic acid and 1 g l−1 vanillin were 0.009 h−1, 0.07 h−1 and 0.08 h−1 with cells obtained at 0%, 12% and 23% (v/v) hydrolysate respectively (Fig. 1B). Thus, as expected, the specific growth rates in the presence of 10 g l−1 acetic acid and 1 g l1 vanillin were lower than in the presence of 5 g l−1 acetic acid and 1 g l−1 vanillin. As predicted, the specific growth rate was always higher when the cells were propagated in the presence of 23% (v/v) hydrolysate than with 12% and 0% hydrolysate. Furthermore, the lag phase was longer in cells propagated without any hydrolysate (0% v/v) than with 12% and 23% hydrolysate, irrespective of the acetic acid concentration.
Mira and colleagues (2010) showed that after subjecting S. cerevisiae cells to a certain level of acetic acid, growth arrest occurred but cell growth was resumed after a lag phase. However, when these pre-adapted cells were used to re-inoculate medium under the same conditions with same acetic acid concentration, no delay in cell growth was observed what would suggest that some changes at genomic level occur during the adaptation. In a similar way, our results also showed that cells pre-grown in the presence of acetic acid – as was the case for cells propagated with 12% and 23% hydrolysate – were more able to tolerate acetic acid (Fig. 1).
Fermentation experiments
To study the differences in ethanol production and xylose fermentation capacity according to the propagation strategy, cells propagated with different concentration of hydrolysate were used to ferment wheat straw hydrolysate diluted to 50% (v/v). This concentration was fixed according to previous experiments to have an inhibitory hydrolysate without compromising the fermentability of the medium.
After propagation, cells were harvested by centrifugation at 5000 r.p.m. for 5 min at room temperature, and the cell pellet was weighed and diluted with sterile water to obtain the desired inoculum size for the fermentation experiments (1.5 g dry weight of cells l−1).
When 0% (v/v) hydrolysate was added during the propagation, no xylose was consumed in the following fermentation step (Fig. 2A). On the other hand, 40% and 98% of the xylose was consumed when 12% and 23% (v/v) hydrolysate, respectively, was added during the propagation (Fig. 2B and C). Also, the glucose fermentation capacity was affected by the addition of hydrolysate during the propagation because in the case where there was no hydrolysate addition, the wheat hydrolysate was very inhibitory to the yeast and only 4 g l−1 of glucose was consumed in 115 h. Glucose, however, was depleted in less than 24 h when 12% or 23% (v/v) hydrolysate was used during the propagation. The improved xylose and glucose co-fermentation capacity in hydrolysate-propagated cells was translated into an increase in ethanol yield from total sugars, from 0.24 to 0.32 g g−1, when 12% and 23% (v/v) hydrolysate, respectively, was used in the propagation step.
Figure 2.
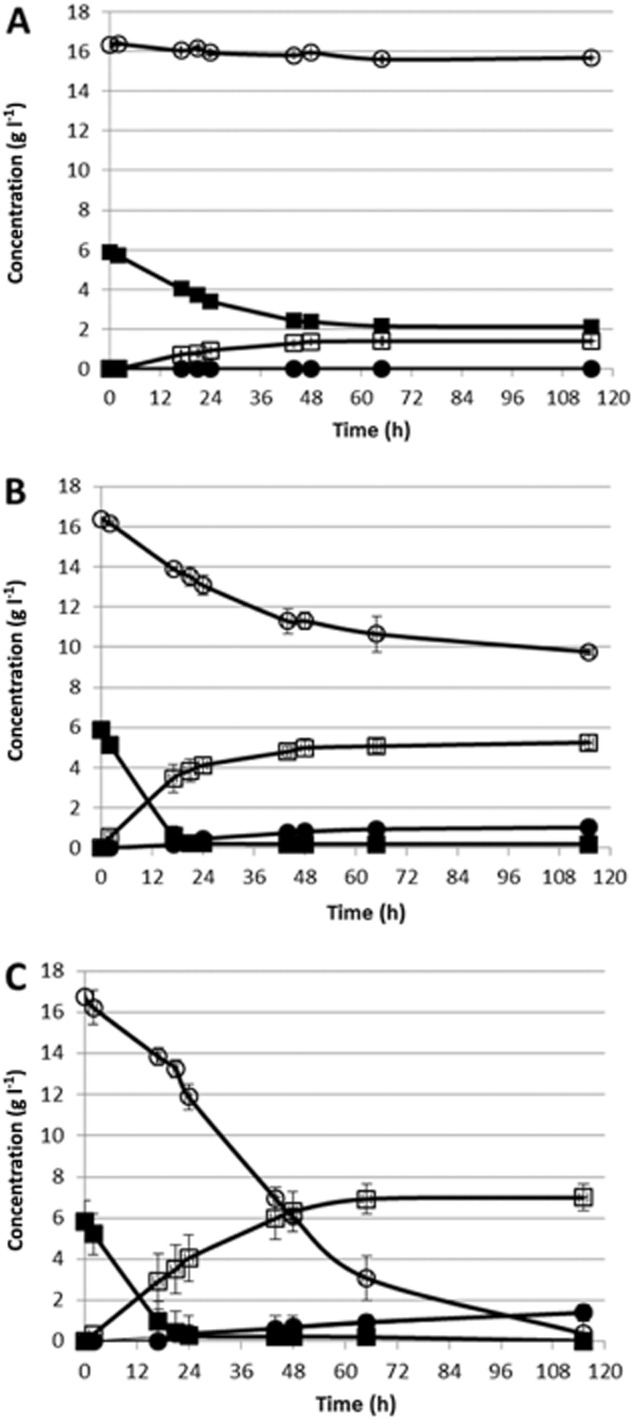
(○) Xylose, (●) xylitol, (■) glucose and (□) ethanol levels in fermentation of 50% (v/v) diluted wheat straw hydrolysate. Experiments were performed for 115 h at 30°C and pH 5.5, in 150 ml semi-anaerobic shake flasks using glycerol traps to allow CO2 outflow and no inflow of oxygen. Di-ammonium phosphate at 0.5 g l−1 was used as nitrogen source. The inoculum size was fixed at 1.5 g l−1 dry weight. Hydrolysate added during the propagation step: (A) 0% (v/v), (B) 12% (v/v) and (C) 23% (v/v).
As discussed above, the XR/XDH pathway has different co-factor preferences for the two enzymes – (NAD(P)H in the case of XR and NAD+ in the case of XDH – which leads to redox imbalance and xylitol production at the expense of ethanol production.
Advantageously, when cells were grown in 23% (v/v) hydrolysate, the xylitol yield was as low as 0.08 g g−1 despite the high amount of xylose consumed, as compared with 0.15 g g−1 when 12% (v/v) hydrolysate was used in the propagation. This could be due to an improved capacity for furfural detoxification acquired during propagation with hydrolysate; it is known that furfural can act as an external electron acceptor, re-oxidizing NADH and reducing xylitol secretion (Ask et al., 2013). However, further experiments would be necessary to understand this interesting effect.
These results clearly show that choosing a propagation strategy that leads to more robust and better-adapted cells leads to drastically improved fermentation performance.
Quantitative polymerase chain reaction
For quantitative polymerase chain reaction (qPCR) analysis, a cell suspension (2 ml) from propagation cultures was centrifuged at 4000 r.p.m. for 3 min, washed with 0.9% NaCl and centrifuged again. The pellet was frozen in liquid nitrogen and stored at −80°C until RNA extraction.
The expression of genes related to tolerance to inhibitors and stress resistance, i.e. ADH6, ALD6, CTA1 and ZWF1, was studied in cells propagated under different conditions.
The TAF10 gene was used as internal reference gene. It was found to have stable expression in all samples because its Ct value did not vary significantly. The primer sequences used in the analysis were designed from the sequences listed in the Saccharomyces Genome Database (http://www.yeastgenome.org/) and stated in Table 1. Furthermore, the relative quantification of gene expression was evaluated using the comparative ΔΔCt method (Livak and Schmittgen, 2001).
Table 1.
Primer sequences used in the qPCR analysis
| Gene | Forward primer | Reverse primer |
|---|---|---|
| ADH6 | GTCTTGGTGGTATCGGCAGTATGGGTA | ATGTCGGTAAGGGAGGAAGCACAGACTA |
| ALD6 | ACCCAAGAGAAAGAGGCCGTCTACTAAG | GCTCTAAGGTGGTGAAGTTCATGTAGCC |
| CTA1 | CAGTACGGTAAATCTGAGGACGGGTCT | GACCGCTTTGTACTGCAGTCTGATCTC |
| ZWF1 | GACATTACTGATATCTGCGGGTCTGCT | GGGAACTTGGAAGGGTCTCTGATAAAG |
| TAF10 | TACCCGAATTTACAAGAAAAGATAAGA | ATTTCTGAGTAGCAAGTGCTAAAAGTC |
ADH6 encodes an alcohol dehydrogenase, and ALD6 encodes an aldehyde dehydrogenase. Both enzymes are involved in resistance to phenolic fermentation inhibitors (Petersson et al., 2006; Park et al., 2011). It has even been shown that yeast clones that overexpress ADH6 have increased capacities for reducing furfural and HMF (Almeida et al., 2009). As indicated in Fig. 3, the expression of ADH6 was upregulated when the cells were grown in the presence of hydrolysate. There was a 1.2-fold and a 1.8-fold difference when they were propagated on 12% and 23% (v/v) hydrolysate respectively. According to the fermentation results, the upregulation of this gene could indicate an improvement in the ethanol production process in the presence of inhibitors.
Figure 3.
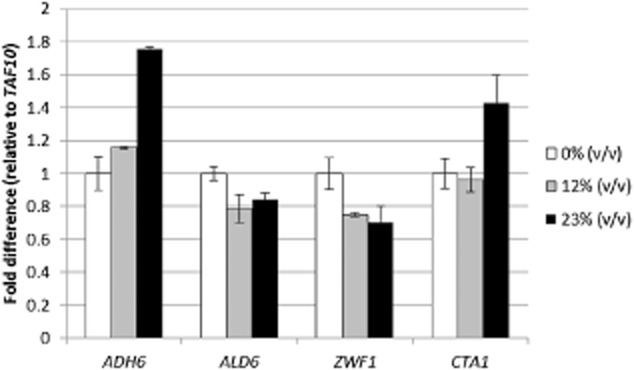
The RNA was extracted using an RNeasy kit (Qiagen) with DNase treatment, according to the manufacturer's protocol. The samples were subjected to reverse transcription and the cDNA was then used for qPCR. Expression of ADH6, ALD6, ZWF1 and CTA1 relative to TAF10 in S. cerevisiae. Expression of these genes related to resistance to inhibitors and to the stress response was quantified using Brilliant® II SYBRGreen QPCR Master Mix, forward and reverse primers (0.5 μM each) and 2 μl cDNA. The qPCR procedure was performed using a Stratagene Mx3005P instrument. The qPCR program started with an initial denaturation for 10 min at 95°C. Amplification consisted of 40 cycles of 30 s at 95°C and 1 min at 65°C. This was followed by 1 min at 72°C for elongation of the amplicons.
When the cells were grown in the presence of wheat straw hydrolysate, no significant differences were observed in ALD6 expression (Fig. 3). However, this gene has been found to be upregulated during xylose fermentation in mineral media in the presence of HMF and furfural (Ask et al., 2013).
ZWF1 encodes a cytoplasmic glucose-6-phosphate dehydrogenase that catalyses the first step in the pentose phosphate pathway, and it is also associated with sensitivity to furfural and HMF. Disruption of ZWF1 has been found to increase the ethanol yield and reduce the xylitol yield in a xylose-fermenting recombinant strain of S. cerevisiae (Jeppsson et al., 2002). These findings were attributed to an altered flux through the pentose phosphate pathway and may explain the highest xylose fermentation ability observed in cells propagated with 12% (v/v) and 23% (v/v) hydrolysate when ZWF1 expression was downregulated (Fig. 3).
CTA1 encodes a catalase, which is induced under oxidative stress (Kim et al., 2006). Furthermore, it has recently been shown to be linked to tolerance to furfural and HMF (Kim and Hahn, 2013). Although no differences in CTA1 expression were detected in cells grown at 0% (v/v) or 12% (v/v) hydrolysate, this gene was clearly upregulated when cells were propagated in 23% (v/v) hydrolysate, which may indicate that addition of hydrolysate during propagation also triggers the oxidative stress response.
The qPCR results clearly show that the propagation strategy followed led to changes in stress associated genes. However, the relationship between the observed changes in gene expression and ethanol production will require further research to be fully understood.
Conclusions
Hydrolysate addition during the propagation step pre-adapts the cells to the inhibitors, leading to cells with improved specific growth rates and reduced lag phases in minimal medium with vanillin and acetic acid. Furthermore, the propagation strategy also had a huge effect on the performance of the cells in ethanol production from wheat straw hydrolysates increasing the final ethanol production yields by 80%.
Cells propagated according to an appropriate strategy also show high xylose consumption rates, high ethanol production rates and low xylitol yields.
Depending on the propagation strategy, changes can be found in gene expression and thereby highlight the importance of optimizing the propagation step for achieving efficient ethanol production.
Acknowledgments
We thank Taurus Energy AB for kindly providing the yeast strain KE6-12, and SEKAB for pretreating the wheat straw and supplying the hydrolysate.
Conflict of interest
Lisbeth Olsson does consultancy work for Taurus Energy AB, Lund, Sweden.
References
- Almeida JRM, Bertilsson M, Hahn-Hägerdal B, Lidén G. Gorwa-Grauslund MF. Carbon fluxes of xylose-consuming Saccharomyces cerevisiae strains are affected differently by NADH and NADPH usage in HMF reduction. Appl Microbiol Biotechnol. 2009;84:751–761. doi: 10.1007/s00253-009-2053-1. [DOI] [PubMed] [Google Scholar]
- Alvira P, Tomás-Pejó E, Ballesteros M. Negro MJ. Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: a review. Bioresour Technol. 2010;101:4851–4861. doi: 10.1016/j.biortech.2009.11.093. [DOI] [PubMed] [Google Scholar]
- Ask M, Bettiga M, Duraiswamy VR. Olsson L. Pulsed addition of HMF and furfural to batch-grown xylose-utilizing Saccharomyces cerevisiae results in different physiological responses in glucose and xylose consumption phase. Biotechnol Biofuel. 2013;6:181. doi: 10.1186/1754-6834-6-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Pastor R, Pérez-Torrado R, Garre E. Matallana E. Recent advances in yeast biomass production. In: Matovic D, editor; Biomass – Detection, Production and Usage. Rijeka, Croatia: InTech Publisher; 2011. pp. 201–222. [Google Scholar]
- Gonçalves DL, Matsushika A, de Sales BB, Goshima T, Bon EPS. Stambuk BU. Xylose and xylose/glucose co-fermentation by recombinant Saccharomyces cerevisiae strains expressing individual hexose transporters. Enzyme Microb Technol. 2014;63:13–20. doi: 10.1016/j.enzmictec.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Jeppsson M, Johansson B, Hahn-Hägerdal B. Gorwa-Grauslund MF. Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl Environ Microbiol. 2002;68:1604–1609. doi: 10.1128/AEM.68.4.1604-1609.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Hahn JS. Roles of the Yap1 transcription factor and antioxidants in Saccharomyces cerevisiae's tolerance to furfural and 5-hydroxymethylfurfural, which function as thiol-reactive electrophiles generating oxidative stress. Appl Environ Microbiol. 2013;79:5069–5077. doi: 10.1128/AEM.00643-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Yun H, Iwahashi H. Jin I. Genome-wide expression analyses of adaptive response against medadione-induced oxidative stress in Saccharomyces cerevisiae KNU5377. Process Biochem. 2006;41:2305–2313. [Google Scholar]
- Kim SR, Park YC, Jin YS. Seo JH. Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol Adv. 2013;31:851–861. doi: 10.1016/j.biotechadv.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Klinke HB, Thomsen AB. Ahring BK. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol. 2004;66:10–26. doi: 10.1007/s00253-004-1642-2. [DOI] [PubMed] [Google Scholar]
- Koppram R, Albers E. Olsson L. Evolutionary engineering strategies to enhance tolerance of xylose utilizing recombinant yeast to inhibitors derived from spruce biomass. Biotechnol Biofuel. 2012;5:32. doi: 10.1186/1754-6834-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppram R, Nielsen F, Albers E, Lambert A, Wännström S, Welin L, et al. Simultaneous saccharification and co-fermentation for bioethanol production using corncobs at lab, PDU and demo scales. Biotechnol Biofuel. 2013;6:2. doi: 10.1186/1754-6834-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martín C, Marcet M, Almazán O. Jönsson LJ. Adaptation of a recombinant xylose-utilizing Saccharomyces cerevisiae strain to a sugarcane bagasse hydrolysate with high content of fermentation inhibitors. Bioresour Technol. 2007;98:1767–1773. doi: 10.1016/j.biortech.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Mira NP, Becker JD. Sá-Correia I. Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS A J of Integr Biol. 2010;14:587–601. doi: 10.1089/omi.2010.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno AD, Tomás-Pejó E, Ibarra D, Ballesteros M. Olsson L. Fed-batch SSCF using steam-exploded wheat straw at high dry matter consistencies and a xylose-fermenting Saccharomyces cerevisiae strain: effect of laccase supplementation. Biotechnol Biofuel. 2013;6:160. doi: 10.1186/1754-6834-6-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TTM, Iwaki A, Ohya Y. Izawa S. Vanillin causes the activation of Yap1 and mitochondrial fragmentation in Saccharomyces cerevisiae. J Biosci Bioeng. 2014;117:33–38. doi: 10.1016/j.jbiosc.2013.06.008. [DOI] [PubMed] [Google Scholar]
- Okuda N, Ninomiya K, Takao M, Katakura Y. Shioya S. Microaeration enhances productivity of bioethanol from hydrolysate of waste house wood using ethanologenic Escherichia coli KO11. J Biosci Bioeng. 2007;103:350–357. doi: 10.1263/jbb.103.350. [DOI] [PubMed] [Google Scholar]
- Park SE, Koo HM, Park YK, Park SM, Park JC, Lee OK, et al. Expression of aldehyde dehydrogenase 6 reduces inhibitory effect of furan derivatives on cell growth and ethanol production in Saccharomyces cerevisiae. Bioresour Technol. 2011;102:6033–6038. doi: 10.1016/j.biortech.2011.02.101. [DOI] [PubMed] [Google Scholar]
- Petersson A, Almeida JRM, Modig T, Karhumaa K, Hahn-Hägerdal B, Gorwa-Grauslund MF. Lidén G. A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast. 2006;23:455–464. doi: 10.1002/yea.1370. [DOI] [PubMed] [Google Scholar]
- Tao H, Gonzalez R, Martinez A, Rodriguez M, Ingram LO, Preston JF. Shanmugam KT. Engineering a homo-ethanol pathway in Escherichia coli: increased glycolytic flux and levels of expression of glycolytic genes during xylose fermentation. J Bacteriol. 2001;183:2979–2988. doi: 10.1128/JB.183.10.2979-2988.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás-Pejó E. Fermenting microorganisms for 2nd generation bioethanol production. In: Min-Tze L, editor. Bioprocess Science and Technology. New York, USA: Nova publishers; 2011. pp. 107–208. [Google Scholar]
- Tomás-Pejó E, Ballesteros M, Oliva JM. Olsson L. Adaptation of the xylose fermenting yeast Saccharomyces cerevisiae F12 for improving ethanol production in different fed-batch SSF processes. J Ind Microbiol Biotechnol. 2010;37:1211–1220. doi: 10.1007/s10295-010-0768-8. [DOI] [PubMed] [Google Scholar]
- Tomás-Pejó E, Bonander N. Olsson L. Industrial yeasts strains for biorefinery solutions: constructing and selecting efficient barcoded xylose fermenting strains for ethanol. Biofuel Bioprod Bior. 2014;8:626–634. [Google Scholar]
- Verduyn C, Postma E, Scheffers WA. Van Dijken JP. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol. 1990;136:395–403. doi: 10.1099/00221287-136-3-395. [DOI] [PubMed] [Google Scholar]


