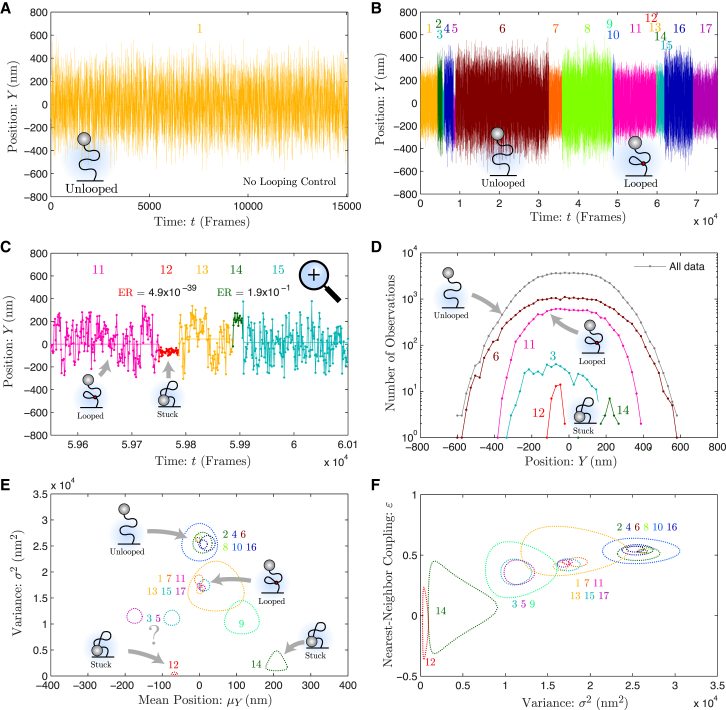
Figure 2.
Analysis of experimental TPM data: protein-induced DNA looping measured by TPM. (A) Position trace for the no-looping control. The y position of the bead shown for 1.5 × 104 frames. In the absence of protein-induced looping, only a single state is identified by change-point analysis, corresponding to the unlooped configuration. (B) Position trace for protein-induced DNA looping. The y position of the bead shown for 7.5 × 104 frames. Seventeen states were identified by change-point analysis. The trace is colored by state and the state number is shown above the trace. A representative example of the unlooped and looped state is shown. (C) High-resolution time trace. At t ≈ 5.98 × 104 frames, a high-time-resolution trace is shown, which reveals two short-lived states, states 12 and 14. The ER for each of these states is shown. The statistical evidence for state 12 is extremely strong whereas the evidence for state 14 is marginal. (D) Histogram of bead position by state. The histogram for all data and selected states is shown. States 6 and 11 are representative of the unlooped and looped states, respectively. Neither state is well approximated by a Gaussian distribution, as demonstrated by the flatness of the peak of the probability density functions. (E) Mean position and variance by state. The 95% confidence region is shown for each state. The states cluster into two clearly identifiable groups corresponding to the unlooped (2, 4, 6, 8, 10, and 16) and looped (1, 7, 11, 13, 15, and 17) states. In addition to these clusters, there are low-mobility and moderate-mobility states with mean positions offset from zero. The short-lived states with low mobility correspond to sticking events (12 and 14). (F) Variance and nearest-neighbor coupling by state. The 95% confidence region is shown for each state. Again, the states form clusters analogous to (E). For states 12 and 14, ε is approximately zero, consistent with bead sticking. To see this figure in color, go online.