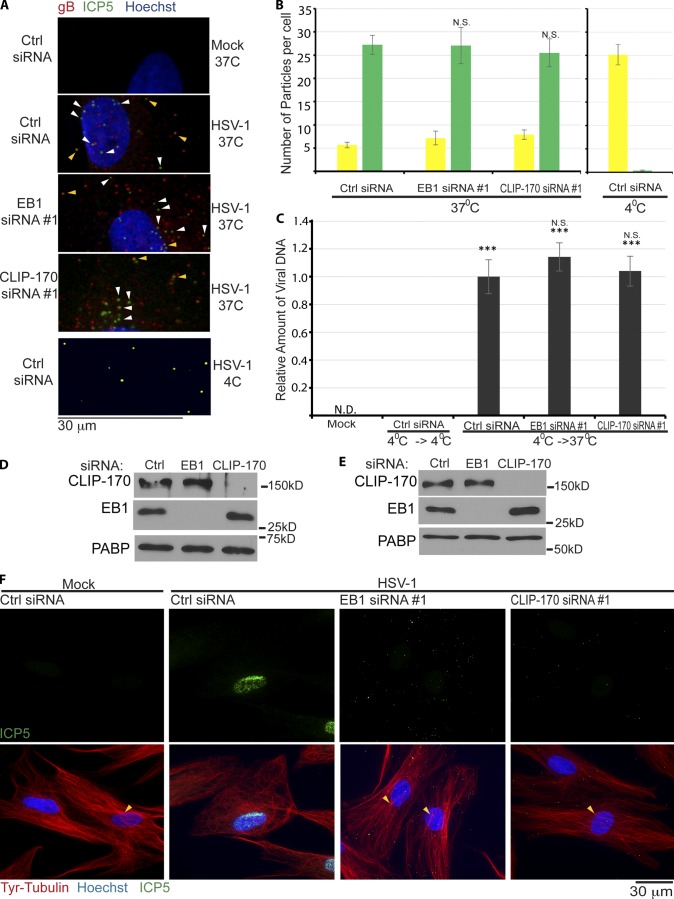
Figure 3.
EB1 and CLIP-170 mediate early post-entry infection. (A) NHDFs were treated with control, EB1, or CLIP-170 siRNAs and infected at MOI 30 for 1 h. Samples were stained for viral glycoprotein, gB (red), or capsid protein, ICP5 (green). Nuclei were stained with Hoechst (blue). Particles lacking envelope (green) or with envelope (yellow) are indicated with white or yellow arrowheads, respectively. 4°C controls illustrate detection of unfused virus particles (yellow). Images were acquired by confocal microscopy, and representative planes are shown. (B) Random fields of view from samples in A were used to quantify unfused (yellow) and fused (green) particles. (Side) Control samples infected at 4°C. n = 18 cells with 18–45 particles per cell. Error bars represent standard error of the mean. Statistical analysis demonstrated no significant differences between control and EB1 or CLIP-170 siRNA–treated samples (one-way ANOVA; n = 18/group; F(2,51) = 0.096; P = 0.909). (C) Virus entry assay. NHDFs were mock infected or infected at MOI 30 at 4°C. Cultures were either maintained at 4°C (4°C→4°C) as a control for unfused virus, or shifted to 37°C (4°C→37°C) to allow virus entry. After removal of unpenetrated virus, viral DNA levels in cell lysates were measured by qRT-PCR. Error bars represent standard error of the mean. ***, P < 0.0001 versus control group at 4°C (one-way ANOVA; n = 3/group; F(3,8) = 98.588; P < 0.0001). Post-hoc analysis revealed levels of viral DNA in infected cells were unaffected by depletion of either EB1 (P = 0.377) or CLIP-170 (P = 0.960). (D) WB analysis of samples from cells that underwent 4°C→37°C shifting as described in C, confirming siRNA target depletion in each case. (E and F) NHDFs were treated with control, nontargeting siRNA, or siRNAs targeting either EB1 or CLIP-170. Cultures were then mock infected or infected at MOI 20 for 4 h. (E) WB analysis confirmed siRNA target depletion. (F) Fixed samples were stained for Tyr-tubulin (red) or the capsid protein ICP5 (green). Nuclei were stained with Hoechst (blue). Representative widefield images are shown. Arrowheads point to areas characteristic of centrosomal nucleation readily observed in either uninfected cells or infected cells depleted of EB1 or CLIP-170, but not detectable in control siRNA–treated cells infected with HSV-1. N.D., not detectable.