Abstract
Formation of secondary walls is a complex process that requires the coordinated and developmentally regulated expression of secondary wall biosynthetic genes. In Arabidopsis thaliana, a transcriptional network orchestrates the biosynthesis and deposition of the main SCW components in xylem and fiber cells. It was recently reported that interacting TALE homeodomain proteins BEL-LIKE HOMEODOMAIN6 (BLH6) and KNOTTED ARABIDOPSIS THALIANA7 (KNAT7) negatively regulate secondary cell wall formation in the interfascicular fibers of Arabidopsis inflorescence stems. Members of the Arabidopsis OVATE FAMILY PROTEIN (OFP) family of transcriptional regulators have been shown to physically interact in yeast with various KNAT and BLH proteins, forming a proposed TALE-OFP protein interaction network. This study presents molecular and genetic data indicating that OFP1 and OFP4, previously reported to interact with TALE homeodomain proteins, enhance the repression activity of BLH6, supporting a role for these OFPs as components of a putative multi-protein transcription regulatory complex containing BLH6 and KNAT7.
Keywords: gene regulation, interfascicular fiber, protein complex, secondary cell wall, xylem
At certain stages of plant development, the synthesis of secondary cell walls is the major metabolic activity, for example, during inflorescence stem development in the model plant Arabidopsis thaliana (Arabidopsis)1 and wood formation in trees.2 Secondary cell walls are major carbon sinks in developing plants, and secondary cell wall formation is precisely controlled by a complex network of transcription factors that function in specialized cells in stems, roots, anthers, and siliques.3-9 Members of this transcriptional network, comprising a set of transcription factors with strong feed-forward character, have been identified by molecular and genetic analyses in various model plant species (Arabidopsis, Oryza, Populus),8,10-14 as well as in Zinnia and Arabidopsis cell trans-differentiation systems.15-17 The currently understood SCW transcriptional network is composed of several families of transcription factors (TFs) including NAC domain, MYB, zinc-finger proteins and homeodomain proteins. A large-scale yeast one-hybrid study has identified putative target genes of many of these TFs, enhancing our understanding of the regulatory network.18 The TALE homeodomain protein KNOTTED ARABIDOPIS THALIANA7 (KNAT7) is a negative regulator of secondary cell wall biosynthesis in Arabidopsis interfascicular fibers (IF).19 We recently showed that KNAT7 interacts with the TALE homeodomain protein BELL-LIKE HOMEODOMAIN6 (BLH6) and that the KNAT7-BLH6 complex operates as a repression module in IF secondary cell wall formation.20 knat7 and blh6 mutants also show a display irregular xylem (irx) phenotypes of collapsed xylem vessel cells,19,20 suggesting potentially different regulatory roles in secondary cell wall formation in these cells.20
Arabidopsis OVATE FAMILY PROTEINS (OFPs) are a family of 18 transcriptional regulators characterized by a conserved 70-aa OVATE domain but lacking recognizable DNA binding domains.21,22 Most OPFs are transcriptional repressors, and several Arabidopsis OFPs were shown to interact with TALE homeodomain proteins in a yeast 2-hybrid screen, suggesting a TALE homeodomain-OPF interaction network.22 The interactions between KNAT7 and OFP4 and OFP1 have been further characterized in vivo and in vitro,23 and the interaction of either OFP1 or OFP4 with KNAT7 was demonstrated to enhance KNAT7s repression activity.23 The ofp4 mutant and ofp1ofp4 double mutant display irx phenotypes similar to those found in knat7 and blh6, and the OFP4 overexpression phenotype is suppressed in a knat7 mutant background, suggesting that a KNAT7-OFP4 complex is required both for aspects of secondary wall formation and for other developmental processes. Recently, we demonstrated a physical interaction between BLH6 and KNAT7 in vivo and in vitro.20 BLH6 also functions as a transcriptional repressor. When transiently expressed together, interaction of BLH6 with KNAT7 enhanced the repression activity of BLH6 or KNAT7 expressed alone.20 Furthermore, KNAT7 and BLH6 appear to work together to repress IF secondary cell wall deposition at least partially by repressing REVOLUTA expression.20 However, the potential roles played by OFP1 and OFP4 in the KNAT7-BLH6 repression complex has not been investigated.
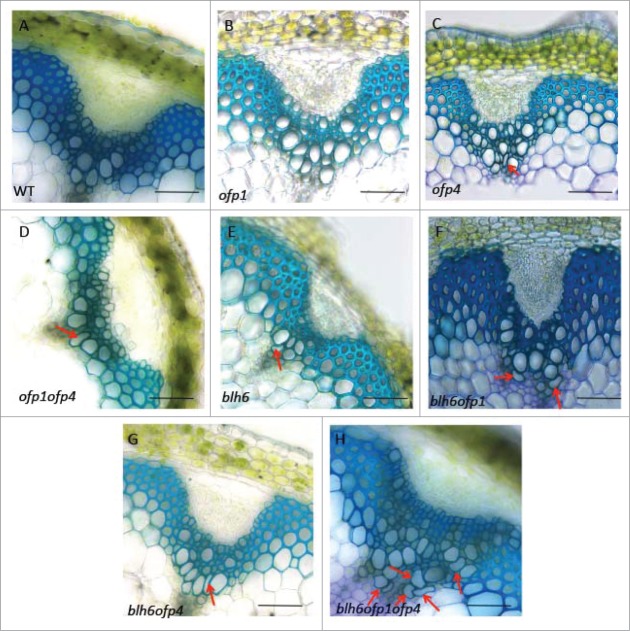
Initial data from 3-hybrid experiments confirmed potential protein-protein interactions between OFP1 and BLH6, between OFP4 and BLH6, and between OFP1, BLH6 and KNAT7 (data not shown). To examine the functional outcomes of BLH6-OFP interactions in planta, we generated blh6 ofp1, blh6 ofp4 and blh6 ofp1 ofp4 plants by crossing an ofp1, ofp4 or ofp1 ofp4 mutants with blh6. Double mutants and triple mutants identified in the F2 populations had no obvious morphological differences compared with wild type. Anatomy at the bases of inflorescence stems taken from 6-week-old plants was analyzed by staining with toluidine blue (Fig. 1). A mild irx phenotype was observed in sections from the bases of inflorescence stems of ofp4 and ofp1ofp4 mutants (Fig. 1C and D), while ofp1 appeared similar to wild type (Fig. 1B) as previously described.23 No differences in fiber cell wall thickness were observed in ofp1, ofp4 and ofp1ofp4 mutants (Table 1). Sections from the bases of blh6 inflorescence stems showed a collapsed vessel (irx) phenotype and thicker IF secondary cell walls as previously described (Fig. 1E). In addition, the mild irx phenotypes observed in the vessels and thicker IF secondary cell wall phenotypes of blh6 ofp1 and blh6 ofp4 mutants were similar to the single blh6 mutant phenotype (Fig. 1F and 2; Table 1). However, the blh6 ofp1ofp4 mutant had an enhanced irx phenotype relative to blh6, and similar IF secondary cell wall thickness to blh6 (Fig. 1H and 2; Table 1).
Figure 1.
Inflorescence stem secondary cell wall phenotypes of blh6 ofp1, blh6 ofp4 and blh6 ofp1ofp4.Stem cross sections were taken from 3 cm from the bases of the inflorescence stems of 6-week-old plants, stained with toluidine blue and examined for alterations in anatomy and secondary wall thickness. (A–D) Representative cross-sections from a wild type (WT, A) control, ofp1 (B), ofp4 (C) and ofp1ofp4 (D) stem showing vascular bundles and interfascicular fibers. (E-H) Representative cross-section from a blh6 (E), blh6 ofp1 (F), blh6 ofp4 (G) and blh6 ofp1 ofp4 (H) stem showing vascular bundles and interfascicular fibers with thicker secondary cell walls relative to wild type. Bars, 50 µm. Arrows indicate the irx vessels.
Table 1.
Interfascicular fiber cell wall thickness in wild-type and mutant plants
| Genotype | Cell wall thickness1 |
|---|---|
| Wild-type | 1.69 ± 0.44 |
| blh6 | 2.72 ± 0.322 |
| ofp1 | 1.54 ± 0.36 |
| ofp4 | 1.79 ± 0.37 |
| blh6 ofp1 | 2.87 ± 0.562 |
| blh6 ofp4 | 2.93 ± 0.412 |
| blh6 ofp1 ofp4 | 2.91 ± 0.452 |
Mean cell wall thickness in µm ± SD; n = 50 cells.
Significantly different from wild type, P < 0.01.
Figure 2.
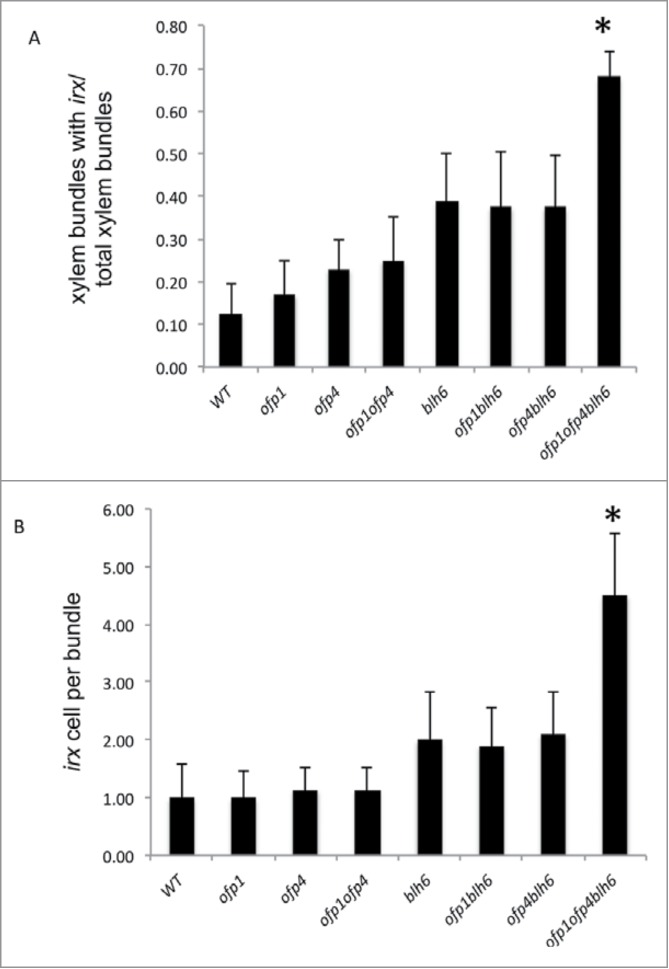
Quantification of the irx phenotype in vascular bundles of the basal internode of 6-week-old stems of wild-type and mutant plants. (A) The irx phenotype was quantified by calculating the number of vascular bundles with vessels showing an irx phenotype per total number of vascular bundles examined. (B) The irx phenotype was quantified by calculating the number of vessels showing an irx phenotype per vascular bundle exhibiting an irx phenotype. n = 24 vascular bundles examine per genotype. Asterisks indicate significant differences from blh6 based on Student's t-tests.
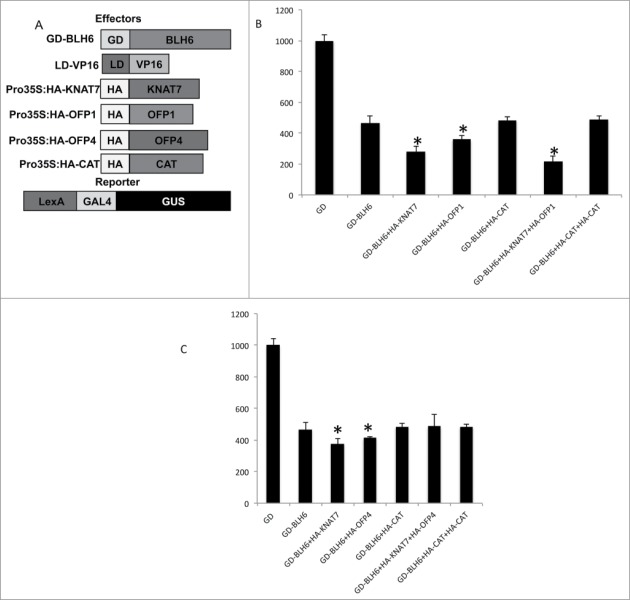
Previous studies showed that OFP1 and OFP4 function as transcriptional repressors in protoplast transfection assays.21-24 To test if the interaction between BLH6 and OFP1 or OFP4 affects BLH6 transcriptional activity similarly to their enhancement of KNAT7 repression activity,23 we employed a protoplast transfection system24 (Fig. 3). We used one set of trans-activating effector plasmids containing GD fusions to BLH6 (GD-BLH6), a second trans-activating effector plasmid containing a Pro35S:LexA DB (LD) fused to VP16 (LD-VP16), and a third effector with OFP1, OFP4 or KNAT7 under the control of the CaMV 35S promoter with an N-terminal hemagglutinin (HA) tag (Pro35S:HA-OFP1; Pro35S:HA-OFP4; Pro35S:HA-KNAT7; Fig. 3A). Reporter plasmids contained LexA-Gal4:GUS, in which LexA and GAL4 DB binding sites were placed upstream of the GUS gene. These experiments were carried out in protoplasts derived from knat7 mutant leaf mesophyll cells, to avoid complications arising from the potential presence of endogenous KNAT7.
Figure 3.
Test of the transcriptional activities of different BLH6-KNAT7-OFP complexes. (A) Effectors and reporter constructs used in the transfection assays. (B) GUS activity in protoplasts derived from Arabidopsis rosette leaves of knat7 loss-of-function mutant plants co-transfected with an LD-VP16 transactivator plasmid and different combinations of effector plasmids (HA-KNAT7, HA-CAT,HA-OFP1 and GD-BLH6) together with the reporter plasmid LexA-GAL4:GUS. Error bars represent the standard deviations, n = 3 transfections from one representative biological replicate. (C) GUS activity in protoplasts derived from Arabidopsis rosette leaves of knat7 loss-of-function mutant plants co-transfected with an LD-VP16 transactivator plasmid and different combinations of effector plasmids (HA-KNAT7, HA-CT,HA-OFP4 and GD-BLH6) together with the reporter plasmid LexA-GAL4:GUS. Error bars represent the standard deviations, n = 3 transfections from one representative biological replicate. 10 µg of an HA-CAT (chloramphenicol acetyltransferase) plasmid was used in place of the HA-KNAT7, HA-OFP1 or HA-OFP4 effector to keep the amount of effector plasmid DNA introduced into protoplasts constant. Asterisks indicate significant differences from GD-BLH6 based on Student's t-tests. Significantly different from GD-BLH6, P < 0.05.
Figure 3B and C show that co-transfection of HA-KNAT7 with GD-BLH6 reduced LexA-Gal4:GUS reporter gene activity relative to GD-BLH6 alone, as previously demonstrated.20 HA-OFP1 or HA-OFP4 co-transfected with GD-BHL6 and LexA-VP16 repressed LexA-Gal4:GUS reporter gene activity relative to GD-BLH6 repression activity alone. Figure 3B also indicates that co-transfection of both HA-OFP1 and HA-KNAT7 with GD-BHL6 further enhanced BHL6 repression activity; i.e. it reduced LexA-Gal4:GUS reporter gene activity relative to transfection of GD-BLH6 and HA-KNAT7 or GD-BLH6 and HA-OFP1 alone. However, in contrast, the combination of HA-OFP4, HA-KNAT7 and GD-BLH6 had no additive effect in reducing reporter gene expression relative to co-transfection of GD-BLH6 and HA-KNAT7 or GD-BLH6 and HA-OFP4 (Fig. 3C). This result was consistent between 3 independent protoplast transfection assays, supporting a model in which OFP1, BLH6 and KNAT7 can form a transcriptional complex that functions to repress VP16 transcriptional activity. However, OFP4 appears to function differently in its interaction with BLH6 and KNAT7.
The inability of HA-OPF4 to enhance the repression activity of the interacting GD-BLH6 and HA-KNAT7 proteins suggests that a functional OFP4-BLH6-KNAT7 complex may not form upon co-transfection. This is consistent with our data from yeast 3-hybrid assays (data not shown) indicating that KNAT7 seems to interfere with the interaction between BLH6 and OFP4, since BLH6 interacts with OFP4 only in the absence of KNAT7 according to these assays. This is consistent with the observation that OFP4, like BLH6, interacts with the KNAT7 Meinox domain20,23 while OFP1 does not.23 This suggests that OFP4 and BLH6 may compete for a common KNAT7 interaction domain, preventing formation of an OFP4-BLH6-KNAT7 complex. In contrast, since KNAT7 interaction domains for OFP1 and BLH6 are different,20,23 an OFP4-BLH6-KNAT7 complex capable of enhanced transcriptional repression (Fig. 3B) may efficiently form.
OFP4–KNAT7 and KNAT7-BLH6 complexes have been shown to influence secondary wall formation in Arabidopsis.20,23 Several lines of evidence from this study suggest a connection between OFP proteins and the BLH6-KNAT7 transcription repression complex that regulates secondary wall formation.20 A protoplast transfection system used to determine the effects of the interaction of KNAT7, OFPs and KNAT7 + OFPs with BLH6 on BLH6 transcriptional activity (Fig. 3) demonstrated that KNAT7 and OFP1 work together in a complex with BLH6 to further enhance BLH6 transcriptional repression, reducing the levels of VP16-activated reporter gene expression observed in the presence of GD-BLH6 alone. These data suggest that BLH6 and BLH6-KNAT7 transcriptional repression is enhanced by formation of a larger complex containing OFP1. However, comparisons of the inflorescence stem phenotypes of blh6, ofp1 and ofp4 single mutants relative to wild type revealed that thicker IF secondary cell walls are only found in blh6 and that the IF secondary cell wall phenotypes are similar among blh6 ofp1, blh6 ofp4 and blh6 ofp1 ofp4 mutants. This may indicate that OFP1 and OFP4 have redundant functions with each other and/or with other OFPs that mask loss-of-function phenotypes, or that neither OFP1 nor OFP4 plays a strong role in IF secondary wall formation.
On the other hand, the blh6 ofp4 double mutant displays mild irx phenotypes similar to that of the blh6 and ofp4 single mutants, supporting the hypothesis that BLH6 and OFP4 work as a complex involved in control of cell wall deposition in xylary vessels, in which case the loss of function of either one would lead to a defective complex. Interestingly, the blh6 ofp1 ofp4 triple mutant exhibited a noticeably enhanced irx phenotype relative to blh6 ofp1 and blh6 ofp4, demonstrating that OFP1 and OFP4 together could enhance BLH6 function in regulating secondary cell wall formation in xylary vessels, and also suggesting functional redundancy between OFP1 and OFP4. The enhanced irx phenotype of blh6 ofp1 ofp4 relative to the single mutants is similar to that observed in the blh6 knat7 mutant, relative to those single mutants.20 These additive effects suggest that multiple TF complexes containing BLH6, KNAT7, OFP1/4, and possibly other TALE homeodomain and OFP protein combinations could exist, which control different aspects of vessel element SCW biosynthesis and deposition. Based on genetic data, it has been proposed that OFP5 negatively modulates the activity of a BLH1 - KNAT3 regulatory complex during embryo sac development in the Arabidopsis.25 By analogy, our findings suggest that OFP1 and OFP4 may modulate the activity of the BLH6 - KNAT7 complex in regulating secondary wall formation in certain cell types or at specific developmental stages.
Funding
Support for this research is from a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant to CJD, and by the NSERC CREATE grant “Working on Walls.”
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
We thank the ABRC and The Arabidopsis Information Resource (TAIR; Carnegie Institute of Washington, Stanford, CA, USA) for seeds and biological information. We also thank Shucai Wang (Northeast Normal University, China) providing vectors for protoplast transfection assays.
References
- 1.Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 1997; 9:689-701; PMID:9165747; http://dx.doi.org/ 10.1105/tpc.9.5.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plomion C, Leprovost G, Stokes A. Wood formation in trees. Plant Physiol 2001; 127:1513-23; PMID:11743096; http://dx.doi.org/ 10.1104/pp.010816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong R, Lee C, Ye Z-H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci 2010; 15:625-32; PMID:20833576; http://dx.doi.org/ 10.1016/j.tplants.2010.08.007 [DOI] [PubMed] [Google Scholar]
- 4.Demura T, Ye ZH. Regulation of plant biomass production. Curr Opin Plant Biol 2010; 13:299-304; PMID:20381410; http://dx.doi.org/ 10.1016/j.pbi.2010.03.002 [DOI] [PubMed] [Google Scholar]
- 5.Wang HZ, Dixon RA. On-Off switches for secondary cell wall biosynthesis. Mol Plant 2012; 5:297-303; PMID:22138968; http://dx.doi.org/ 10.1093/mp/ssr098 [DOI] [PubMed] [Google Scholar]
- 6.Hussey SG, Mizrachi E, Creux NM, Myburg AA. Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front Plant Sci 2013; 4:325; PMID:24009617; http://dx.doi.org/ 10.3389/fpls.2013.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuetz M, Smith R, Ellis B. Xylem tissue specification, patterning, and differentiation mechanisms. J Exp Bot 2013; 64:11-31; PMID:23162114; http://dx.doi.org/ 10.1093/jxb/ers287 [DOI] [PubMed] [Google Scholar]
- 8.Ko JH, Jeon H-W, Kim WC, Kim JY, Han KH. The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann Bot 2014; 114:1099-107; PMID:24984711; http://dx.doi.org/ 10.1093/aob/mcu126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong R, Ye ZH. Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 2015; 56:195-214; PMID:25294860; http://dx.doi.org/ 10.1093/pcp/pcu140 [DOI] [PubMed] [Google Scholar]
- 10.Zhong R, Lee C, Ye ZH. Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol 2010; 152:1044-55; PMID:19965968; http://dx.doi.org/ 10.1104/pp.109.148270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 2005; 19:1855-60; PMID:16103214; http://dx.doi.org/ 10.1101/gad.1331305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant EH, Fujino T, Beers EP, Brunner AM. Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 2010; 232:337-52; PMID:20458494; http://dx.doi.org/ 10.1007/s00425-010-1181-2 [DOI] [PubMed] [Google Scholar]
- 13.Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008; 20:2763-82; PMID:18952777; http://dx.doi.org/ 10.1105/tpc.108.061325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong R, Lee C, Ye ZH. Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol 2010; 152:1044-55; PMID:19965968; http://dx.doi.org/ 10.1104/pp.109.148270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda H. Tracheary element differentiation. Plant Cell 1997; 9:1147-56; PMID:12237380; http://dx.doi.org/ 10.1105/tpc.9.7.1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts K, McCann MC. Xylogenesis: the birth of a corpse. Curr Opin Plant Biol 2000; 3:517-22; PMID:11074384; http://dx.doi.org/ 10.1016/S1369-5266(00)00122-9 [DOI] [PubMed] [Google Scholar]
- 17.Oda Y, Mimura T, Hasezawa S. Regulation of secondary cell wall development by cortical microtubules during tracheary element differentiation in Arabidopsis cell suspensions. Plant Physiol 2005; 137:1027-36; PMID:15709154; http://dx.doi.org/ 10.1104/pp.104.052613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor-Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, et al.. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015; 517:571-5; PMID:25533953; http://dx.doi.org/ 10.1038/nature14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li E, Bhargava A, Qiang W, Friedmann MC, Forneris N, Savidge RA, Johnson LA, Mansfield SD, Ellis BE, Douglas CJ. The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol 2012; 194:102-15; PMID:22236040; http://dx.doi.org/ 10.1111/j.1469-8137.2011.04016.x [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, You S, Taylor-Teeples M, Li WL, Schuetz M, Brady SM, Douglas CJ. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 2014; 26:4843-61; PMID:25490916; http://dx.doi.org/ 10.1105/tpc.114.128322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Chang Y, Guo J, Zeng Q, Ellis BE, Chen J-G. Arabidopsis ovate family proteins, a novel transcriptional repressor family, control multiple aspects of plant growth and development. PLoS One 2011; 6:e23896; PMID:21886836; http://dx.doi.org/ 10.1371/journal.pone.0023896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hackbusch J, Richter K, Müller J, Salamini F, Uhrig JF. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci USA 2005; 102:4908-12; PMID:15781858; http://dx.doi.org/ 10.1073/pnas.0501181102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li E, Wang S, Liu Y, Chen J-G, Douglas CJ. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J 2011; 67:328-41; PMID:21457372; http://dx.doi.org/ 10.1111/j.1365-313X.2011.04595.x [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Chang Y, Guo J, Chen J-G. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J 2007; 50:858-72; PMID:17461792; http://dx.doi.org/ 10.1111/j.1365-313X.2007.03096.x [DOI] [PubMed] [Google Scholar]
- 25.Pagnussat GC, Yu H-J, Sundaresan V. Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 2007; 19:3578-92; PMID:18055603; http://dx.doi.org/ 10.1105/tpc.107.054890 [DOI] [PMC free article] [PubMed] [Google Scholar]