Figure 4.
The Distance Unit Provides an Internal Template that Varies on a Daily Basis
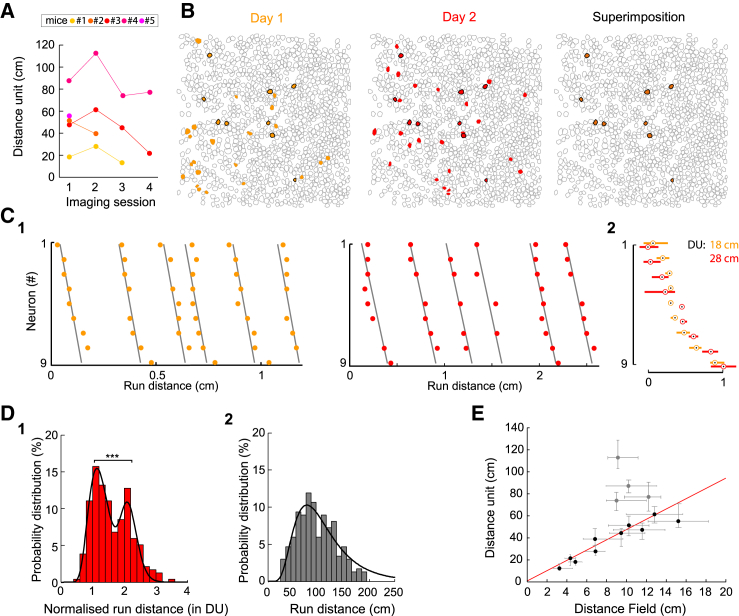
(A) Evolution of the “distance unit” over four daily imaging sessions for five mice (colored lines).
(B) Representative example of the contour maps of imaged cells indicating neurons involved in distance sequences on the first day where sequences could be imaged (yellow, left) and the next (middle, red) or both (orange, right and black contours on all three maps).
(C) (C1) Rasterplots showing the activation, as a function of distance, of the nine cells involved in distance sequences on both days. Neurons are ordered based on their average activation delay on the first day; day 1 (left), day 2 (right). (C2) Superimposed average rasterplot of the sequences on both days (day 1, orange; day 2, red); the median delay of onset (dot) and the corresponding interquartile range (rectangle) are indicated for each cell and calculated taking cell #5 as reference. Note that the “distance unit” (DU) calculated for each day (same color code) varied significantly.
(D) Probability distribution histograms of the normalized (1) and absolute (2) run distance (n = 235 run epochs, 10 sessions with a median run distance smaller than 2 DU). The normalized run distance displays a bimodal distribution (r2 = 0.98) with a first peak at 1.1. DU and a significant second one at 2.1 DU (Figures S4D–S4H, ∗∗∗p < 0.001). The absolute distance histogram distribution is log-normal (r2 = 0.91) with a peak at 77 cm.
(E) Graph indicating the correlation between the average size of single-cell distance fields (median values and interquartile ranges, see Experimental Procedures) and the “distance unit”; the fit (red, Pearson, R = 0,94, p < 0.001) indicates that the average distance field represents one-fifth of the distance unit.