Figure 2.
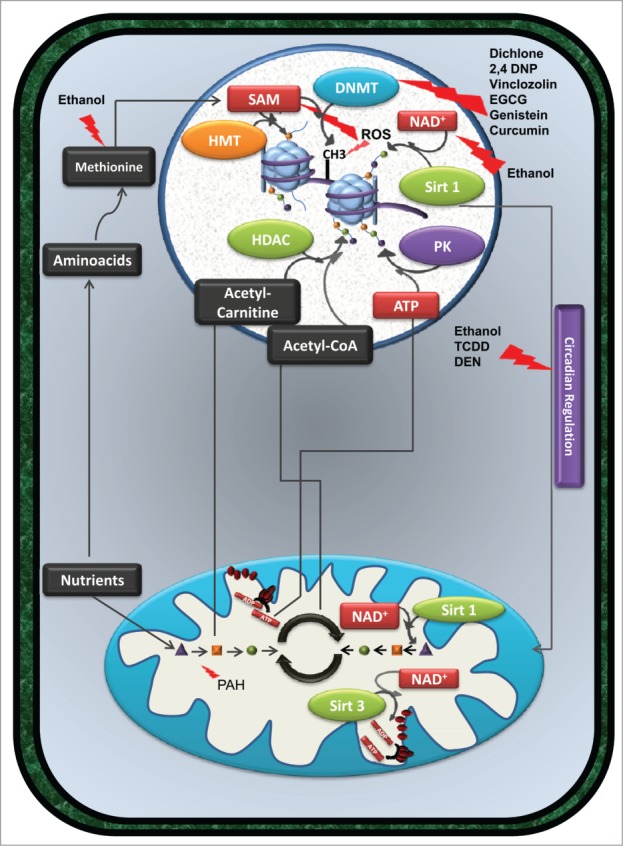
(See previous page). Exposure to xenobiotic compounds does not always cause cell death, but can induce persistent alterations in gene expression and/or metabolism. By interfering directly with the activity of specific enzymes or modulating the availability of specific metabolites, certain xenobiotics promote cellular epigenetic remodeling. Compounds such as dichlone, 2,4 DNP, vinclozolin, EGCG, genistein, and curcumin affect the activity of DNMT enzymes, altering DNA methylation patterns. Also, ethanol consumption, by interfering with methionine metabolism and consequently with SAM levels, results in a similar effect. Furthermore, the detoxification of ethanol by alcohol dehydrogenases decreases the availability of NAD+ and consequently influences the activity of HDAC such as sirtuin 1, inducing alterations in histone acetylation (green dots connected to histones in the figure). Alterations in the cellular redox balance also influence epigenetic regulation of gene expression. In fact, oxidizing conditions induced by certain xenobiotics inhibit the activity of SAM synthetase and consequently decrease the availability of SAM, necessary for DNMT and HMT activity. ROS produced by several compounds promotes the oxidation of DNA bases; a mutagenic event may interfere with DNA methylation. Interplay between epigenome, metabolism and circadian clocks can also be observed. Since the cellular response to xenobiotics can be regulated by the Clock gene system, compounds such as ethanol, TCDD, PCBs, and DEN alter the expression of clock gene expression and affect mitochondrial metabolism and consequently epigenetics. Metabolism altering agents, such as PAH, can limit the availability of certain metabolites, such as acetyl-carnitine, Acetyl-CoA or ATP, consequently affecting histone acetylation and phosphorylation (purple dots linked to histones in the figure) levels. HDAC enzymes such as Sirt1 and Sirt3 not only promote histone deacetylation but also promote deacetylation of several mitochondrial enzymes involved in mitochondrial metabolism and oxidative phosphorylation. Alterations in the acetylation status of those enzymes affect metabolism and metabolites availability, affecting epigenetic remodeling. These alterations, if occurring in developmental key points, can be passed to downstream generations, increasing their susceptibility to diseases. Abbreviations in figure: 2,4 DNP - 2,4 dichlorophenol; ADP - Adenosine diphosphate; ATP - Adenosine triphosphate; DEN – diethylnitrosamine; DMNT - DNA methyltransferases; EGCG - epigallocatechin-3-gallate; HDAC - histone deacetylases; HMT – histone methyltransferases; NAD+ - Nicotinamide adenine dinucleotide (oxidized form); PAH - polycyclic aromatic hydrocarbons; PK – protein kinase; SAM - s-adenosyl-L-methionine; Sirt1 - Sirtuin 1; Sirt3 – Sirtuin 3; TCDD - 2,3,7,8-tetrachlorodibenzo-p-dioxin, PCB – polychlorinated biphenyls.
