Figure 5.
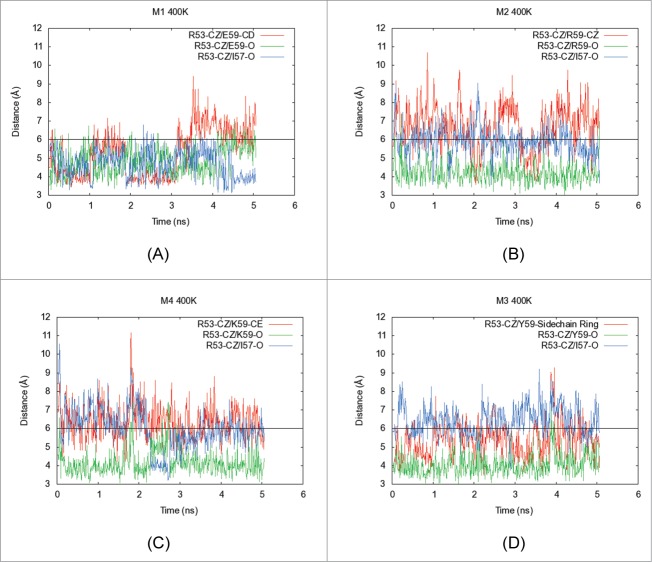
Five nanoseconds-long molecular dynamics (MD) simulations were performed on the Fv portions of (A) M1, (B) M2, (C) M4 and (D) M3 at elevated temperatures (400 K). In each plot, X-axis shows the simulation time (ns) and Y-axis indicates distances (Å) between specific atoms of interacting residues in the structural context for position 59 in the light chain of MAB 1. The side chain and backbone distances are shown for interactions formed by the residue at position 59 and I57 with the side chain functional group of R53. The red line shows the distance between central atom in side chain of residue at position 59 and the Cξ atom in the side chain of R53. The green line indicates the distance of the backbone carbonyl atom of the residue at position 59 with the Cξ atom in the side chain of R53. Similarly, the blue line indicates the distance of the backbone carbonyl atom of I57 from the Cξ atom in the side chain of R53. When the distance between carbon atoms is below 6 Å, an interaction is considered to be formed (black horizontal line). This figure shows that side chain guanidium group of R53 engages in promiscuous salt bridging interactions with the back bone carbonyl of I57, and the backbone carbonyl as well as side chain carboxylate of E59 in the parent mAb, M1. Upon charge reversal at position 59 (M2 and M4 variants), the electrostatic repulsions between the side chains of the residue at position 59 and R53 weaken this electrostatic network leading to destabilization of the Fab as evidenced by decreased Tm2 values for M2 and M4. However, the charge neutralization variant, M3, restores the network among these 3 residues by forming tyrosine ring π interactions with the guanidium group of R53. Analogous simulations were also performed at 300 K. These are shown in the supplementary material (Fig. S2A–S2D).