Figure 1.
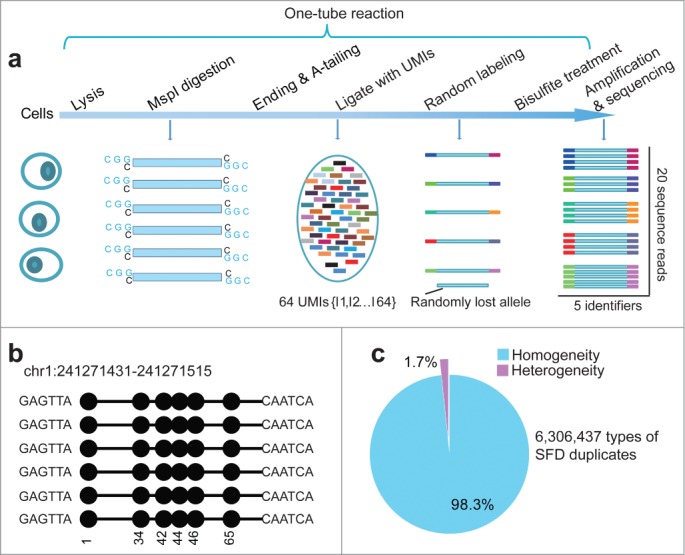
Signatures of quantitative reduced representation bisulfite sequencing (Q-RRBS). (A) A schematic of the Q-RRBS method used for starting materials from either a trace amount of cells or single cells. Briefly, we first performed cell lysis, MspI digestion, repairing/A-tailing, UMI-adapter ligation, and bisulfite treatment in single-tube reactions, and then performed amplification, sequencing, and deduplicated analysis. Adapters containing UMIs (unique molecular identifiers) at their 3′ ends are represented by the short, variously colored boxes within the ellipse. The random-labeling step showed that both ends of each double-stranded DNA molecule randomly ligated with one of the 26 distinct UMI adapters. After deduplicated analysis, the 5 identifiers indicated that only 5 molecules were present, whereas the original analysis showed that 20 sequencing reads were present. (B) An example of single-molecular-fragment-derived (SFD) duplicates that contained identical UMIs (6-bp sequences located at both ends of black lines) and aligned to the same position of the genome. This type of SFD duplicates contained 6 fragments that displayed homogeneity of the DNA methylation pattern (same distribution of methylated CpGs in the sequencing reads). Filled circles represent methylated CpGs. (C) Of the 6,306,437 types of SFD duplicates that derived from 6,306,437 positions of the diploid genome, 98.3% displayed homogeneity of the DNA methylation pattern (shown by the example in Fig. 1b) in each type of the SFD duplicates.
