Abstract
Evidences suggest that tumor microenvironment may play an important role in cancer drug resistance. Sphingosine kinase 2 (SphK2) is proposed to be the key regulator of sphingolipid signaling. This study is aimed to investigate whether the combination of molecular targeting therapy using a specific inhibitor of SphK2 (ABC294640), with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) can enhance the apoptosis of non-small cell lung cancer (NSCLC) cells. Our results revealed that NSCLC cells' sensitivity to TRAIL is correlated with the level of SphK2. Compared with TRAIL alone, the combination therapy enhanced the apoptosis induced by TRAIL, and knockdown of SphK2 by siRNA presented a similar effect. Combination therapy with ABC294640 increased the activity of caspase-3/8 and up-regulated the expression of death receptors (DR). Additional investigations revealed that translocation of DR4/5 to the cell membrane surface was promoted by adding ABC294640. However, expression of anti-apoptosis proteins such as Bcl-2 and IAPs was not significantly modified by this SphK2 inhibitor. Overall, this work demonstrates that SphK2 may contribute to the apoptosis resistance in NSCLC, thus indicating a new therapeutic target for resistant NSCLC cells.
Keywords: death receptor, NSCLC, resistance, sphingosine kinase 2, TRAIL
Abbreviations
- NSCLC
non-small cell lung cancer
- TRAIL
tumor necrosis factor-related apoptosis inducing ligand
- SphK2
sphingosine kinase 2
- DR4
death receptor 4
- DR5
death receptor 5
- ABC294640
3-(4-chlorophenyl)-adamantane-1-carboxylic acid (pyridin-4-ylmethyl) amide
- DISC
death-induced signaling complex
- S1P
sphingosine-1-phosphate
- Cer
ceramide
- Bcl-2
B-cell lymphoma 2
- MTT
(3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide
Introduction
Lung cancer is one of the leading causes of cancer-related morbidity and mortality over the world, and 7five percent of newly diagnosed lung cancers are non-small cell lung cancer (NSCLC).1 When diagnosed, almost 75% cases present advanced (stage III or IV) diseases, the overall 5-year survival rate of which is approximately 1–5%.2 Besides, relapsed disease may occur in patients with stage I or II NSCLC after complete resection.3 For advanced disease and postoperative recurrence, systemic or adjuvant chemotherapy is always necessary. However, despite of the rapid development of biological agents targeting specific cancer-related molecules, not all cancer types are sensitive to these agents and new treatment strategies are urgently needed to be designed.
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a member of the tumor necrosis factor (TNF) family 4,5 which plays a vital role during the process of proliferation, differentiation, and apoptosis in tumor cells.6 TRAIL activates the apoptosis pathway by stimulating TRAIL receptors (TRAIL-R1 and TRAIL-R2, also known as death receptors 4 and 5, respectively) on the target cell surface.7 Another 2 receptors, the TRAIL-R3 (DcR1) and TRAIL-R4 (DcR2), also called “decoy receptors,” cannot trigger the apoptotic cascade since the absence of functional death domains. Once bound with corresponding ligands, DR4 or DR5 recruits the death-induced signaling complex (DISC) components onto the inner surface of plasma membrane, where the caspase-8 become activated.8 Caspase-8 initiate the intrinsic and extrinsic apoptotic pathways by activating downstream caspases, such as caspase-3 or cleavage of the Bcl-2 family member Bid.9 An amplification of apoptotic signals occurs after the activation of death receptors.10 TRAIL could be a potential target not only for its outstanding antitumor activity against a broad range of cancer types but also for its minimal cytotoxity to most of the normal cells and tissues.11,12 It has been reported that TRAIL induces apoptosis in NSCLC cell lines and inhibits the growth of NSCLC xenografts.13,14 Therefore, TRAIL is one of the most promising potential agents for cancer therapeutics.
However, many lung cancer cells lines are resistant to apoptosis induced by TRAIL, which limits the therapeutic applicability of this agent. Some reports suggest that TRAIL resistance may result from a combination of overexpression of Bcl-2 family members and other antiapoptotic proteins, upregulated activity of NF-kB pathway, and deficient expression of death receptors and caspases.15,16 However, the TRAIL resistance phenomena could not be explicitly explained in multiple cancer cells.17 As a signal molecule of upstream events, the effects mediated by death receptor are essentially important for initiating apoptosis. And death receptors expressed on cell surface are exclusively functional.18 Our previous study indicated that certain interventions could restore SW480 and Hep-2R cell line's sensitivity to TRAIL by upregulating the cell surface expression of death receptors.19,20 Therefore, further study is required to unravel the mechanism regulating TRAIL resistant signaling and to identify molecular switches that may set the system into the “apoptosis position.” Previous study has indicated that targeting the sphingosine signaling could help to bring the misguided apoptosis back to the right track. 21
Sphingolipids are both structural and functional components of biological membranes.22 It has been found that their metabolites play pivotal roles in the survival and resistance pathways of numerous solid tumors. Ceramide and sphingosine are associated with growth arrest and apoptosis.23 As a metabolic product of sphingosine, sphingosine-1-phosphate (S1P) is demonstrated to be a pro-survival agent.24 Several enzymes contribute to the regulation process involving these bioactive lipids, the so called “sphingolipid rheostat.”25 Among them, sphingosine kinases (SphK) have emerged as the central players who catalyze the phosphorylation of sphingosine to S1P.26 In human cells, 2 SphK isoenzymes exist.27,28 SphK1 is well known for promoting cell growth, survival, cancer development and resistance to multiply treatments.29,30 In contrast, the roles of SphK2 in cancer cells are not fully understood. There is accumulating evidence demonstrating that SphK2 is frequently upregulated and/or overexpressed in tumor tissues or drug-resistant cancer cells, compared to normal tissues and more sensitive cancer cells.31,32 A large numbers of studies have showed that targeting SphK could restore cancer cells' sensitivity to chemotherapy, however to the best of our knowledge, the effect of SphK2 expression on TRAIL-induced apoptosis is not yet discussed in relevant reports.
In this study, in order to elucidate the role of SphK2 in lung cancer cells resistant to TRAIL, we used the ABC294640, a selective sphingosine kinase-2 inhibitor,33 to examine whether ablation of SphK2 can facilitate apoptosis induced by TRAIL in these cells. We demonstrated that the resistance to TRAIL is associated with the expression of SphK2. Our research found that SphK2 suppression acts cooperatively with TRAIL' effects through up-regulating death receptors' surface expression, suggesting that SphK2 might represent a novel approach for the treatment of NSCLC.
Results
Sphingosine kinase is overexpressed in TRAIL resistant lung cancer cells
A number of studies have tested the different expression of sphingosine kinase between tumor and normal tissues,34,35 but few have examined the difference between chemoresistant cancer and sensitive cancer. In our study, we examined in vitro the anti-proliferative effect of Apo2L/TRAIL in 3 representative human NSCLC cell lines, H460, A549 and H1299 and measured SphK2 expression in order to analyze their correlations. In MTT assays, TRAIL displayed an IC50 value of 125.23ng/ml in H460 cells; in contrast, A549 and H1299 cells were relatively resistant to TRAIL (Fig. 1A). Furthermore, according to the results of real time RT-PCR, both Sphk1 and Sphk2 were overexpressed in TRAIL resistant NSCLC cell lines compared with the TRAIL-sensitive H460 cells, the positive control. In addition, Sphk2 expression was extremely high in the 2 TRAIL-resistant NSCLC cell lines (Fig. 1B). Besides, A549 and H1299 cells also showed a higher SphK2 protein level than H460 cells (Fig. 1C, D). These results suggest that various expression levels of sphingosine kinase, especially Sphk2, may contribute to NSCLC cells' resistance to TRAIL.
Figure 1.
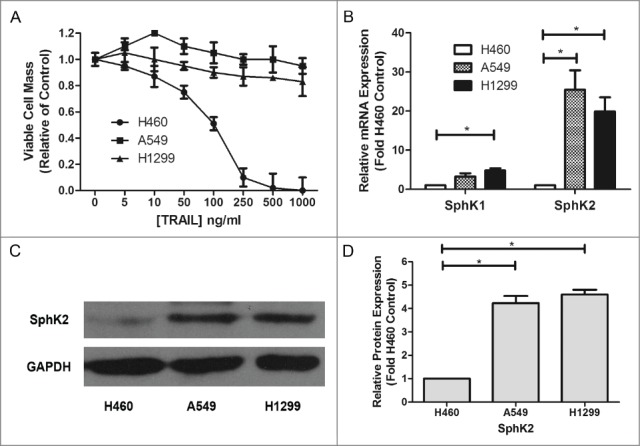
Dysregulation of sphingosine kinases in TRAIL resistant lung cancer cells. (A) H460, A549 and H1299 cells were plated at 1 × 105/ml cells per well in 96-well plate. The following day cells were treated with indicated concentrations of TRAIL for 24 h. Data are presented as percent of vehicle treated samples. Mean values of 5 different experiments (*p < 0.05). (B–D) qRT-PCR analysis and Western blot for expression of sphingosine kinase isoforms in TRAIL resistant lung cancer cells. Data are expressed as fold-change relative to H460 cell control as normalized to internal GAPDH. Data points and error bars represent the mean ±SEM of 3 independent experiments. Columns represent mean density of 3 different experiments (*p < 0.05)
Targeting sphingosine kinase-2 enhances the sensitivity of TRAIL in resistant lung cancer cells
As described above, there are conflicting evidences on role of Sphk2, with several supporting its anti-proliferation effects and others arguing for its pro-proliferation effects. Some argue that the roles of Sphk2 appear to be specific to cell types and cell conditions.36 According to our results, mRNA levels and protein levels of SphK2 in these 2 TRAIL-resistant NSCLC cells were substantially decreased when Sphk2 expression was knocked down by siRNA, as shown in Figure 2A and B. Cells transfected with siNC were defined as control for subsequent knockdown experiments. SphK2-silenced NSCLC cells were treated with different doses of TRAIL for 24 h, and their viability rate measured by MTT assay was much lower as compared with TRAIL alone (Fig. 2C, D), indicating that SphK2 was actually an important target to enhance the sensitivity of TRAIL.
Figure 2.
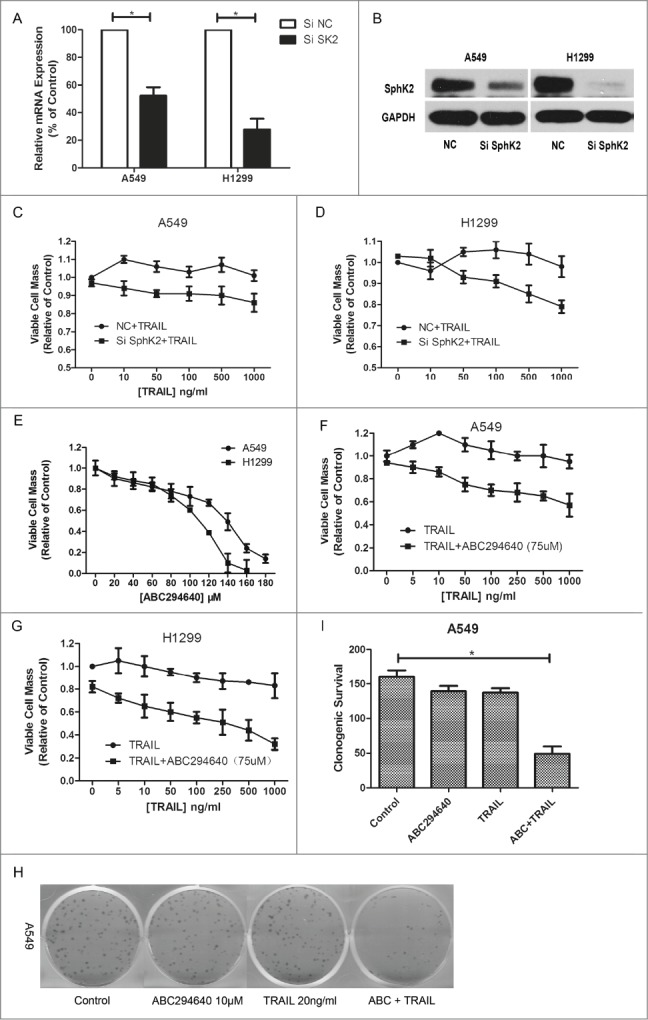
Resensitization of TRAIL-induced cell death by targeting sphingosine kinase 2. (A and B) Cells were transfected with siRNA as indicated, and RT-PCR and Western were carried out after 24 h and 48 h separately to evaluate the efficiency of siSphK2. Data points and error bars represent the mean ±SEM of 3 independent experiments. (C and D) After transfected with siSphK2 for 24 h, A549 and H1299 cells were treated with indicated concentrations of TRAIL for another 24 h. Cell viability was measured by MTT assay. Mean values of 5 different experiments. (E) A549 and H1299 cells were treated with indicated concentrations of ABC294640 for 24 h. Cell viability was measured by MTT assay. Mean values of 5 different experiments. (F and G) Cells were treated with indicated concentrations of TRAIL alone or combined with 75 μM ABC294640 for 24 h. Cell viability was measured by MTT assay. Mean values of 5 different experiments. (H and I) A549 cells were treated with TRAIL (20 ng/ml), ABC294640 (10 μM) or TRAIL+ABC294640 for 10 d Cells were stained with crystal violet and counted under microscope. Colonies ≥30 cells were scored as positive for colony formation. Data are presented as the number of colonies. Columns represent mean data of 3 different experiments (*p < 0.05).
Furthermore, a dose-dependent apoptosis induced by ABC294640, an inhibitor of SphK2, was detected in these 2 lung cancer cell lines (Fig. 2E). In order to determine whether pharmacologic inhibition of Sphk2 could also increase the anti-proliferation of TRAIL, we combined TRAIL and ABC294640 of sublethal dose which would induce less than 20% cell death. After co-treatment for 24h, MTT assay showed that combination treatment promoted cell death both in A549 and H1299 cells, compared with TRAIL alone (Fig. 2F, G). Moreover, in order to imitate the model of clinical patients, we further examined this inhibition effect with clonogenic survival assay which revealed that the combination of TRAIL and ABC294640 led to the elimination of 48% long-term colony formation (Fig. 2H, I). These findings indicated that adding ABC294640 may significantly reduce long term cell survival by restoring cells' sensitivity to TRAIL. Taken together, these results indicate that co-treatment with Sphk2 inhibitors and TRAIL sensitizes the resistant lung cancer cells to apoptosis induced by TRAIL, and combination therapy with these drugs may result in a synergistic biological effect.
Targeting sphingosine kinase-2 enhances apoptosis induced by TRAIL in resistant lung cancer cells
Previous researchers found that several mechanisms may contribute to TRAIL resistance, including lack of caspase-3 and caspase-8 activations, downregulation of its receptors or upregulation of some anti-apoptotic proteins.37,38 Here we examined the effects of TRAIL alone or in combination with ABC294640 on the apoptotic signaling induced by TRAIL.
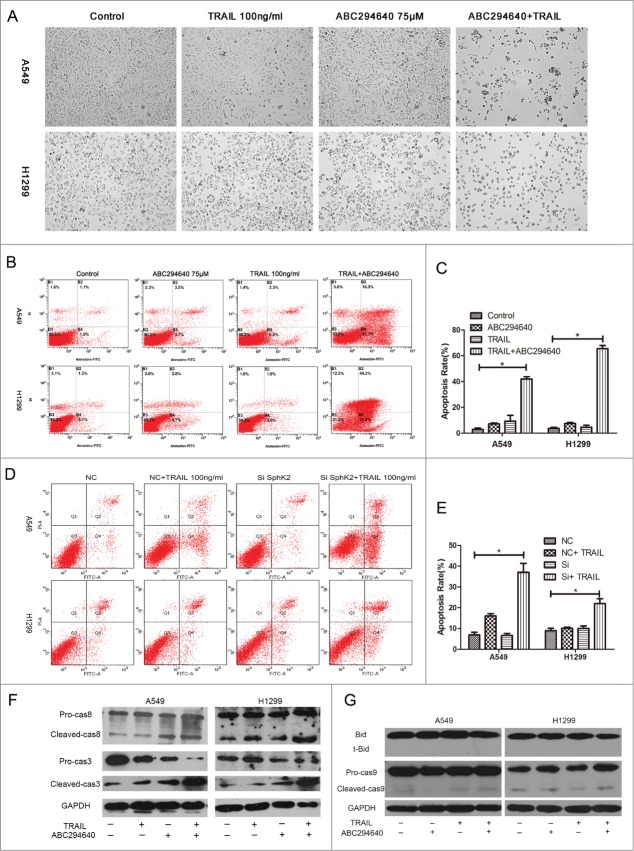
Firstly, we assessed the apoptosis morphologically. Exposed to 75μM ABC294640 for 24 hours, narrowing and rounding was observed in A549 and H1299 cells respectively, with puny influence on cell death. The morphology of TRAIL treated A549 and H1299 cells resembled that of untreated cells, although occasional apoptotic cells were observed. However, combination of ABC294640 and TRAIL caused de-adhesion in virtually all cells and induced apoptosis (Fig. 3A). This is consistent with the results of MTT assay.
Figure 3.
ABC294640 increased TRAIL induced apoptosis. A549 and H1299 cells were grown on 6-well plates to 70-80% confluence and treated with indicated concentrations of TRAIL (100 ng /ml) or ABC294640 (75 mM) or both of the above treatments, or non-treatment. After 24 h exposure, (A) morphologic assessment were captured using Olympus IX70 Inverted Microscope at a magnification of 100, (B-E) cells were stained with PtdIns and Annexin V-FITC, then apoptosis were analyzed by flow cytometry as described in Materials and Methods. Data are presented as total apoptosis rate. Mean §SEM of 3 independent experiments. (F and G) caspase-3/8/9 and Bid were assessed by Western blot of cell lysates, as described in Materials and Methods. GAPDH expression served as a loading control. Representative of 3 experiments.
In addition, the synergistic induction of apoptosis by combination of ABC294640 and TRAIL was confirmed by Annexin V-FITC/PI flow cytometry. The total apoptotic cells were calculated as the sum of percentages of early and late stage apoptotic cells. As shown in Figure 3B and 3C, the combination was associated with a significant increase in the proportion of apoptotic cells compared with single treatment using either ABC294640 or TRAIL in A549 cells. Similar results were observed in H1299 cells with almost 5.42 ± 2.04 (p < 0.05) fold increase in apoptosis induced by TRAIL compared to single treatment. We also examined whether the selective depletion of SphK2 by siRNA has the similar effect as that of ABC294640. Compared with cells transfected with siNC, SphK2-silenced cells showed a higher apoptosis rate when co-treated with TRAIL (Fig. 3D, E).
Then we investigated whether ABC294640 could increase TRAIL -induced caspase cleavage by Western blot. The data showed that combination treatment with ABC294640 and TRAIL strongly stimulated caspase-3 and caspase-8 cleavage in A549 and H1299 cells (Fig. 3F), however cleavage of caspase-9 and Bid was nearly undetectable (Fig. 3G), indicating that the caspase cascade may be involved in the process in which sphingosine kinase 2 inhibitor enhances TRAIL sensitivity, mainly via extrinsic pathways.
Taken together, these results suggest that ABC294640 enhances TRAIL-induced extrinsic apoptosis and cell death.
ABC294640, an sphingosine kinase 2 inhibitor, upregulates the expression of death receptors
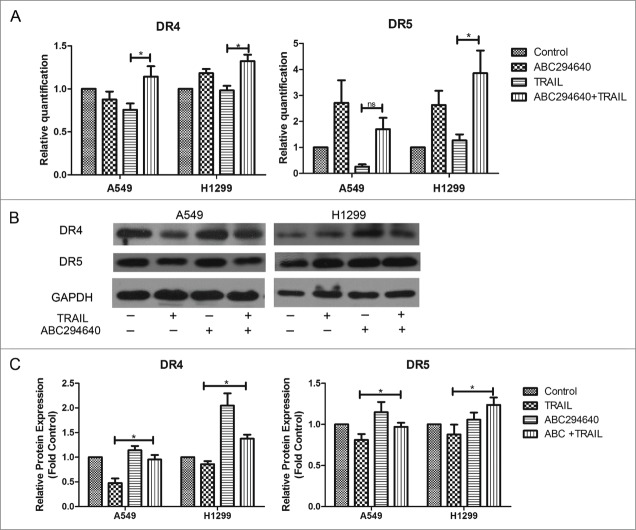
Previous study showed that TRAIL activity was mediated through its interaction with death receptors DR4 and DR5, triggering apoptotic signaling.39 Therefore, to further investigate the mechanism by which ABC294640 enhances the apoptosis induced by TRAIL in NSCLC cells, we examined its effect on the expression of 2 TRAIL death receptors, DR4 and DR5, by RT-PCR and Western blotting. Treating H1299 cells and A549 cells with ABC294640 and TRAIL for 24 hours resulted in slightly increased expression of DR4 and DR5 at the transcriptional level (Fig. 4A). Based on Western blot, it was found that compared with TRAIL alone, ABC294640 alone or combined with TRAIL upregulated DR4 and DR5 expression in both A549 and H1299 cells (Fig. 4B, C). According to these findings, we hypothesize that elevated expression of death receptors might play an important role in sphingosine kinase 2 inhibitor induced restoration of sensitivity to TRAIL.
Figure 4.
ABC294640 upregulated expression of death receptors. (A) Cells were treated with vehicle, TRAIL, ABC294640 or TRAIL+ABC294640 for 12 h and analyzed for mRNA levels of DR4 and DR5 using RT-PCR. Data points and error bars represent the mean±SEM of 3 independent experiments. (B) Expression of DR4 and DR5 in each cells treated with vehicle, TRAIL, ABC294640 or TRAIL+ABC294640 for 12 h were estimated by Western blot. GAPDH expression served as a loading control. Data are expressed as fold-change relative to vehicle control as normalized to internal GAPDH. Representative of 3 experiments. (C) Densitometric analysis of western blot images was performed with Image-Pro Plus 6.0 and data were expressed as fold-change relative to control as normalized to internal GAPDH. P-values shown combined treatment compared to TRAIL alone (*p < 0.05).
ABC294640 promotes the translocation of death receptors from cytoplasm to cell membrane
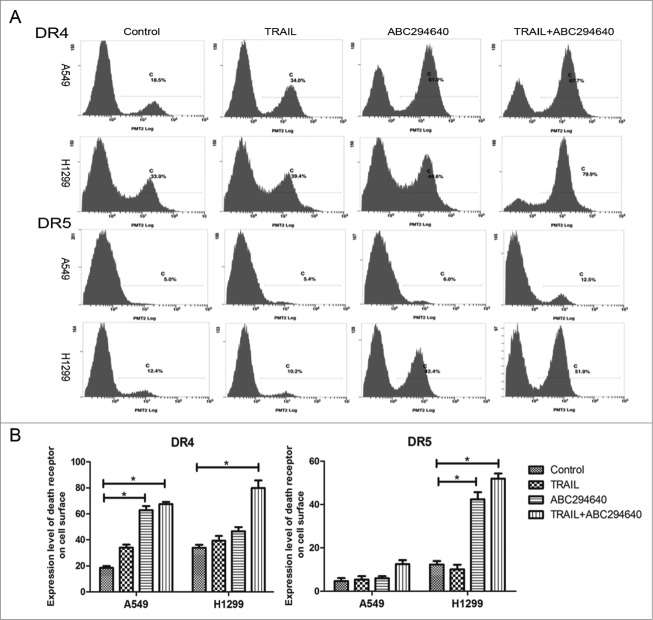
It is well established that only cell membrane surface DR4 and DR5 are able to bind with TRAIL and to transduce apoptotic signals. Accordingly, we performed flow cytometry to analyze the expression of TRAIL death receptors on the cell surface, using FITC/PE-conjugated antibodies. Compared with the negative control (embodied only with second antibody), less expressed DR4 and DR5 was detected on the cell surface in untreated A549 and H1299 cells. 50 μM ABC294640 treatment for 12 hours increased the expression level of death receptors on the cell surface. Furthermore, a significant increase of DR4 and DR5 on the cell membrane surface was found after co-treatment with 50 μM ABC294640 and 100 ng/ml TRAIL, especially in H1299 cells (Fig. 5A, B). Collectively, these results indicate that sphingosine kinase 2 inhibitor up-regulates the cell-surface expression of death receptors that are capable of binding to TRAIL and undergoing apoptotic signal transduction.
Figure 5.
ABC294640 promoted death receptors translocation to cell surficial membrane. (A) The cell surface DR4/DR5 expressions of A549 and H460 cells without any treatment was detected by flow cytometry as described in Materials and Methods. (B) A graphic representation of the mean data presented in (A). Columns represent mean data of 3 different experiments (*p < 0.05).
Discussion
TRAIL is considered as an attractive anticancer agent for its selective toxicity against cancer cells.40 However, low death receptors expression appears to be a major barrier for the intrinsic resistance to TRAIL-based cancer therapy.41 Therefore, identifying novel agents which are capable of restoring TRAIL receptor expression is important to overcome TRAIL resistance. In this study, we verified the hypothesis that resistance to TRAIL may be correlated with the the overexpression of SphK2, the key enzyme which plays a central role in sphingolipide metabolism. This hypothesis was based on previous studies showing (a) SphK2 expression are overexpressed in NSCLC compared with normal lung tissues,31 (b) pharmacological inhibition of SphK2 induces apoptosis and enhances the sensitivity to chemotherapy in breast cancer cells,32 (c) resistance to TRAIL is associated with aberrant sphingolipide signaling and exogenous C6-ceramide can overcome TRAIL resistance in colon cancer cells.21
In this study, statistical differences were detected for SphK2 expression between TRAIL resistant and sensitive NSCLC cells. A549 and H1299 cells that are resistant to TRAIL-induced apoptosis had a higher SphK2 expression level than H460 cells, indicating that sphingolipide signaling may have a connection with TRAIL efficacy. Other studies have also observed specific changes of sphingolipide signaling in drug resistant cancers. For example, the catabolism of glycoceramide (GlcCer) into ceramide (Cer) by nonlysosomal β-glucosidase GBA2 promotes apoptosis in melanoma cells,42 in contrast, the overexpression of GlcCer synthase decreases the endogenous Cer levels, resulting in the development of a multidrug resistance phenotype in many cancer cells.43 Another way through which cancer cells escape from apoptosis associated with Cer accumulation is converting Cer into S1P, which, unlike Cer, acts as a pro-survival signal. Pchejetski et al. found that upregulation of SphK 1/S1P signaling impairs the efficacy of chemotherapy (docetaxel and camptothecin), and pharmacological inhibition or genetic silencing of SphK 1 induce apoptosis in vitro and in vivo.44 However, the knowledge of Sphk 2 in drug resistance is still poorly known. Our study also showed that knockdown of SphK2 did enhance TRAIL-induced cell death. In light of these evidences, our data suggested that Sphk 2 played a pivotal role in TRAIL resistance.
In previous studies, it has been suggested that employment of chemical or genetic approaches specifically targeting sphingolipid dysregulations may be a promising tool for the improvement of TRAIL efficacy. Therefore, we tested whether SphK2-based strategy, by modulating S1P metabolism, was able to enhance TRAIL cytotoxicity and overcome TRAIL resistance in NSCLC cells. ABC294640, a small molecular inhibitor of SphK2 which possesses excellent oral bioavailability and limited toxicity to normal tissues,33 is currently studied in 2 clinical trials for treatment of advanced solid tumors and refractory/relapsed diffuse large B-cell lymphoma (NCT01488513 and NCT02229981). In the present study, we showed that 100ng/ml TRAIL in combination with sublethal dose ABC294640 successfully reduced the viability, proliferation and survival of TRAIL resistant lung cancer cells in all 3 assays. Furthermore, flow cytometric analyses and Western blot analyses revealed that apoptosis induced by TRAIL was enhanced by SphK2 inhibition in A549 cells and H1299 cells. Wafik S. EI-Deiry has reported that regulation of BID cleavage may define if a cell is mitochondria-independent (Type I) or mitochondria–dependent (Types II) in response to TRAIL induced apoptosis.45 Concerning that no truncated Bid was detectable, we considered that A549 and H1299 cells behaved like Type I cells which were mitochondria-independent. Since A549 cells contain wild-type p53 and H1299 cells harbours null p53, our data also suggested that restoration of TRAIL sensitivity by ABC294640 was p53-independent. Consistent with our findings, James W. Antoon's research shows that ABC294640 restores breast cancer chemo-sensitivity through targeting NF-kB.32 Moreover, concomitant administration of ABC294640 and paclitexal to Caov-3 ovarian cancer cells induces apoptotic cell death through activation of caspase-9.46 Taken together, these evidences indicate that targeting SphK2 may be a useful approach to re-sensitize tumors to standard therapy.
ABC294640 was reported to affect a multitude of signaling cascades (ERK, NF-kB, AKT, etc.).47 NF-kB is reported to activate the transcription of anti-apoptotic genes such as c-FLIP, Bcl-xl, Mcl-1 and cIAPs.48,49 Recently, a study concerning TRAIL resistant NSCLC cells found that inhibition of Akt resulted in sensitization for TRAIL apoptosis through dephosphorylation of Akt.50 Blockades in apoptosis can be overcome by combined therapy with targeted agents, conventional chemotherapy and radiotherapy. Generally, these agents restore TRAIL sensitivity by upregulating TRAIL death receptors and/or downregulating anti-apoptotic proteins.51,52 Therefore, in this study, we tried to explain the mechanisms through which ABC294640 sensitized TRAIL on these 2 aspects. We found that despite minor changes in genetic expression level of death receptor, the protein level of DR4 was upregulated following ABC294640 treatment with or without TRAIL. In order to further investigate this change, we examined cell-surface expression level of death receptors. Our data suggested that this molecular inhibitor of SphK2 significantly promoted both DR4 and DR5 surface expression. In a previously published study, the acquired resistance of SW480 cells to TRAIL is associated with significant down-regulation of DR4 on the cell surface; and restoration of TRAIL sensitivity is accompanied by an increase in surface death receptor expression in cancer cells with acquired TRAIL resistance.53 Moreover, we also did some research to evaluate whether the anti-apoptotic proteins' expression was affected by ABC294640. However, expression of apoptosis modulators remained almost unchanged, including Bax, Bcl-2 and IAPs (Fig. S1A–C). Collectively, we hypothesis that ABC294640 enhanced TRAIL induced apoptosis by promoting death receptors' translocation onto the cell surface. However, the underlying mechanism needs further investigation.
As we all know, autophagy is a reversible catabolic adaptive process responsible for the degradation and reutilization of some cell components which promote cell survival. However, existence of autophagsome in moribund cells indicates autophagic cell death. Interestingly, it is reported that ABC294640 could induce autophagy in tumor cells and might be an effective adjuvant to drug-induced tumor cell apoptosis.54 Our unpublished data demonstrated that ABC294640 induced autophagy in a dose-dependent manner in TRAIL-resistant NSCLC cells. Therefore, in subsequent studies we will examine and verify the restoration of TRAIL sensitivity induced by this SphK2 inhibitor in vivo and will quest the role which autophagy might play in the restoration of TRAIL-sensitivity.
Given that TRAIL is an excellent antitumor agent with little toxicity for normal tissue, whereas may not be effective against all cancers, identifying a target that enhances its sensitivity is of increasing importance. In conclusion, our studies demonstrate that pharmacologically targeting sphk2 with orally bioavailable inhibitors has promising potential to improve treatment for TRAIL-resistant lung cancer.
Materials and methods
Reagents
Tumor necrosis factor-related apoptosis (TRAIL), designed for clinical use, was obtained from Shanghai TRAIL Bio-technical Co., Ltd. ABC294640 [3-(4-chlorophenyl)-adamantane-1-carboxylic acid (pyridin-4-ylmethyl)-amide] was provided by Active Biochem. Dimethyl sulfoxide (DMSO) was purchased from Sigma. Chemical Corporation.
Cell culture
Human NSCLC cell lines, i.e. NCI-H1299,A549 and NCI-H460, were provided by the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China).These cells were cultured in RPMI-1640 medium (Hyclone, USA) supplemented with 10%(v/v) of heat-inactivated fetal bovine serum. Cells were maintained at 37°C in a fully humidified atmosphere containing 5% CO2.
siRNA transfection
The SphK2-specific small interfering RNA (siRNA) and negative control were purchased from Shanghai GenePharma Co. Ltd. (Shanghai, China). Sequences for the SphK2 specific siRNA are as follows: SphK2 (pre-designed siRNA ID 1677, sense 5′-GGGUAGUGCCUGAUCAAU Gtt-3′, antisense 5′-CAUUGAUCAGGCACUACCCtc-3′). Cells were plated into 6-well plates without antibiotics for transfection. After 24 h, 4 ul of Lipofectamine 2000 (Invitrogen) and 8ul of siRNA was mixed with 400 ul RPMI-1640 serum free medium for 20min at room temperature. Then it was added to 3ml of antibiotic growth media for each well. The efficiency of transfection was evaluated by rt-PCR and Western blot.
Cell viability assay
Cells were cultured at 1 × 105/ml in 100 μl aliquots in 96-well tissue culture plates (Corning, USA) and allowed to attach overnight. A blank control group (nutritive medium only), a negative control group (untreated cells) were designed in this experiment. Briefly, cells were treated with indicated concentrations of TRAIL (0, 10, 50, 100, 500, 1,000 ng/ml) with or without ABC294640 for 24 h. Following treatment, 20 μl of 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT, 5 mg/ml) reagent was added in each well, and then incubated for 4h. The formazan crystals were solubilized in 100 μl DMSO at 37°C. The absorbance values of the solution in each well were measured at 490 nm using a microplate reader (Beckman, USA). Cell viability was determined by the formula: cell viability (%) = (absorbance of the treated wells-absorbance of the blank control wells)/(absorbance of the negative control wells-absorbance of the blank control wells) × 100%. All MTT experiments were performed in 5 replicates and repeated at least 3 times.
Clonogenic survival assay
Cells were plated in 6-well plates at a density of 500 cells per well in 2 ml of 10% RPMI. After twenty-4 hours, cells were treated with TRAIL and/or ABC294640 and then monitored for colony growth. After ten days, the cells were affixed to the plate with 3% glutaraldehyde. Following fixation for 30 min, the plates were washed and stained with a 0.2% solution of crystal violet and 20% methanol, then washed with PBS and dried. Formations of greater than 30 cells were considered sufficient size for a positive colony count. Results were adjusted to those of the DMSO vehicle control and half maximal inhibitory concentration (IC50) and statistical significance values were calculated using dose-response curves with Graph Pad Prism 5.0 (Graph Pad Software, San Diego, CA USA).
Apoptosis assays
Cells were grown on 6-well plates to 70–80% confluence and treated with indicated concentrations of rhTRAIL or ABC294640 or both of the above treatments, or non-treatment. After 24 h exposure, cells were trypsinized, resuspended in 10% fetal bovine serum-containing media, washed twice in PBS, and resuspended in 400 μl Annexin binding buffer. 5 μl of Annexin-V-FTIC (Bestbio, Shanghai, China) solution was added and the mixture was kept at room temperature for 15 min, then 10 μl of 20 μg/ml propidium iodide (Bestbio, Shanghai, China) for 5 min in the dark at room temperature. Finally, cell apoptotic profile was analyzed immediately in flow cytometer (Beckman, USA). The percentage of apoptotic cells was obtained from a bivariate histogram of Annexin-V-FTIC labeled-cells versus propidium iodide-labeled DNA.
Flow cytometric detection of death receptor surface expression
Procedure for cell surface expression of death receptors DR4 and DR5, following the manufacturer's recommendations. In brief, cells at 70–80% confluence were treated with indicated concentration of TRAIL with or without ABC294640 for 12 h, and then cells were harvested by trysinisation and washed twice in ice-cold PBS. Cells were incubated in 40 μl PBS containing 1% goat serum for 40 min at room temperature. After washed with PBS for 3 times, cells were incubated with anti-DR4 or anti-DR5 rabbit antibody (dilution ratio is 1:50) overnight at 4°C. Afterwards, cells were washed once with PBS containing 1% FBS and incubated with FITC/PE-conjugated goat anti rabbit IgG (dilution ratio is 1:50) for 45 min in dark at room temperature. Finally, cells were washed with PBS and resuspended in 0.5 ml PBS for flow cytometry analysis.
Real time RT-PCR
Total RNA was isolated from different cells using TRIzol reagent (Invitrogen, Carlsbad, CA USA). RT-PCR was carried out using High-Fidelity RT-PCR kit (Invitrogen, Carlsbad, CA) as per the manufacturer's instructions. The concentration of RNA was determined using an ultraviolet spectrophotometer. Reverse transcription (RT) was performed using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). Human18S rRNA and glyceraldehyde–3–phosphate dehydrogenase (GAPDH) were used as endogenous control transcripts for normalization of the target transcripts. Primers for PCR were designed to span intron/exon junctions to minimize amplification of residual genomic DNA. The primer sequences for GAPDH, DR4, DR5, SphK1 and SphK2 are as follows (sense and anti-sense, respectively):
GAPDH:5′-TGGAAGGACTCATGACCACA-3′;
5′-TTCAGCTCAGGGATGACCTTL-3;
DR4: 5′-AGAGAGAAGTCCCTGCACCA-3′;
5′-GTCACTCCAGGGCGTACAAT-3′;
DR5: 5′-CACCAGGTGTGATTCAGGTG-3′;
5′-CCCCACTGTGCTTTGTACCT-3′;
SphK1: 5′-AATGAAGACCTCCTGACCAACTG-3′;
5′-GACGCCGATACTTCTCACTCTCT-3′;
SphK2: 5′-GGAGGAAGCTGTGAAGATGC-3′;
5′-GCAACAGTGAGCAGTTGAGC-3′;
The PCR reaction was carried out as follows: step 1: 95°C 3 min, step 2: for 40 cycles 95°C 20 seconds, 60°C 30 seconds, step 3: 70°C 10 seconds, hold at 4°C. Each reaction tube contained 12.5 μl 2× SYBR Green supermix +0.5 μl ROX + 9 μl nuclease-free water + 2 μl 0.1 μg/μl primer (pair) + 1 μl cDNA (0.2 μg/μl). Genes were amplified in triplicate. The average cycle threshold (Ct) value for each group was determined and normalized by the endogenous control Ct value. Relative gene expression was analyzed using the 2-ΔΔCt method.
Western blot analysis
Levels of caspase-3, caspase-8, DR4, DR5 and GAPDH were determined by Western blotting in cell extracts. Cells were washed 3 times with ice-cold PBS, exposed to recommended volume RIPA lysis buffer (Beyotime, Shanghai, China). After centrifugation at 12,000 rpm for 5 min at 4˚C, the supernatants were removed and diluted in 5× loading buffer (Beyotime, Shanghai), boiled for 5 min and loaded onto a 12.5% SDS-PAGE electrophoresis under denaturing conditions, then transferred to PVDF membranes (Beyotime, Shanghai).The membranes were blocked in 5% non-fat milk in TBST (TBS with 0.1% Tween-20) at room temperature. Then the membranes were incubated overnight with the primary antibodies for GAPDH (Sangon Biotech, Shanghai, China), caspase-3 (Cell Signaling Technology, Beverly, MA), caspase-8 (Cell Signaling Technology, Beverly, MA USA), caspase-9 (Cell Signaling Technology, Beverly, MA USA), Bid (Cell Signaling Technology, Beverly, MA), DR4 (BD PharMingen, San Diego, CA USA), DR5 (BD PharMingen, San Diego, CA USA), IAP1 (Sangon Biotech, Shanghai, China) and IAP2 (Cell Signaling Technology, Beverly, MA). Primary antibodies were purchased from Cell Signaling Technology, Inc. After being washed 3 times with TBST, the membranes were incubated for 60 min with secondary infrared conjugated antibodies diluted in 5% milk-TBST at room temperature, then washed 3 times with TBST again. Finally, immunoreactive protein was examined with an enhanced chemiluminescence detection kit (Biyuntian, Shanghai, China). GAPDH was used to normalize protein levels.
Statistical analysis
Each experiment was proformed repeatedly for at least 3 times. Data were presented as the mean value ± standard deviation (SD). Statistical analysis of IC50 values were calculated using Graph Pad Prism 5.0 (Graph Pad Software), using with the equation: Y = Bottom + (Top-bottom)/1 + 10LogEC50 – X. Combination effects were calculated using the formula : (X1a + X2b)/(X1a + X2b), wherein X1 is the effect of drug 1 at concentration a, and X2 is the effect of drug 2 at concentration b and (X1a + X2b) is the effect of the combination of both drugs at concentrations a and b, respectively.32 Experiments comparing multiple concentrations with the control were tested with one-way ANOVA. All statistical analyses were done using SPSS 18.0 software (SPSS, Chicago, IL USA). A P < 0.05 was considered as statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Xiangyu Meng from the Center for Evidence-based Medicine and Translational Medicine of Wuhan University, for his advice on improving language quality of this manuscript.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This research was supported by grants from the National Natural Science Foundation of China (No. 81372498) and the National Natural Science Foundation of Hubei Province (No. 2013CFA006).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69-90; PMID:21296855; http://dx.doi.org/ 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2.Bulzebruck H, Bopp R, Drings P, Bauer E, Krysa S, Probst G, van Kaick G, Muller KM, Vogt-Moykopf I. New aspects in the staging of lung cancer. Prospective validation of the International Union Against Cancer TNM classification. Cancer 1992; 70:1102-10; PMID:1515985; http://dx.doi.org/ 10.1002/1097-0142(19920901)70:5%3c1102::AID-CNCR2820700514%3e3.0.CO;2-5 [DOI] [PubMed] [Google Scholar]
- 3.Detterbeck FC. Diagnosis and treatment of lung canceran evidence-based guide for the practicing clinician. Philadelphia, W.B. Saunders; c2001; Chapter 4, Early Stage Non-Small Cell Lung Cancer (NSCLC);p.181–190. [Google Scholar]
- 4.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, Et A. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 1995; 3:673-82; PMID:8777713; http://dx.doi.org/ 10.1016/1074-7613(95)90057-8 [DOI] [PubMed] [Google Scholar]
- 5.Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem 1996; 271:12687-90; PMID:8663110; http://dx.doi.org/ 10.1074/jbc.271.22.12687 [DOI] [PubMed] [Google Scholar]
- 6.Krammer PH. The CD95(APO-1/Fas)/CD95L system. Toxicol Lett 1998; 102–103:131-7; PMID:10022244 [DOI] [PubMed] [Google Scholar]
- 7.Ashkenazi A. Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat Rev Drug Discov 2008; 7:1001-12; PMID:18989337; http://dx.doi.org/ 10.1038/nrd2637 [DOI] [PubMed] [Google Scholar]
- 8.Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer 2008; 8:782-98; PMID:18813321; http://dx.doi.org/ 10.1038/nrc2465 [DOI] [PubMed] [Google Scholar]
- 9.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 1998; 94:481-90; PMID:9727491; http://dx.doi.org/ 10.1016/S0092-8674(00)81589-5 [DOI] [PubMed] [Google Scholar]
- 10.Merino D, Lalaoui N, Morizot A, Solary E, Micheau O. TRAIL in cancer therapy: present and future challenges. Expert Opin Ther Targets 2007; 11:1299-314; PMID:17907960 http://dx.doi.org/ 10.1517/14728222.11.10.1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacFarlane M. TRAIL-induced signalling and apoptosis. Toxicol Lett 2003; 139:89-97; PMID:12628743; http://dx.doi.org/ 10.1016/S0378-4274(02)00422-8 [DOI] [PubMed] [Google Scholar]
- 12.Wang S, El-Deiry WS. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene 2003; 22:8628-33; PMID:14634624; http://dx.doi.org/ 10.1038/sj.onc.1207232 [DOI] [PubMed] [Google Scholar]
- 13.Bhojani MS, Rossu BD, Rehemtulla A. TRAIL and anti-tumor responses. Cancer Biol Ther 2003; 2:S71-8; PMID:14508083; http://dx.doi.org/ 10.4161/cbt.205 [DOI] [PubMed] [Google Scholar]
- 14.Hao C, Song JH, Hsi B, Lewis J, Song DK, Petruk KC, Tyrrell DL, Kneteman NM. TRAIL inhibits tumor growth but is nontoxic to human hepatocytes in chimeric mice. Cancer Res 2004; 64:8502-6; PMID:15574753; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-2599 [DOI] [PubMed] [Google Scholar]
- 15.Takeda K, Stagg J, Yagita H, Okumura K, Smyth MJ. Targeting death-inducing receptors in cancer therapy. Oncogene 2007; 26:3745-57; PMID:17530027; http://dx.doi.org/ 10.1038/sj.onc.1210374 [DOI] [PubMed] [Google Scholar]
- 16.Kim YS, Schwabe RF, Qian T, Lemasters JJ, Brenner DA. TRAIL-mediated apoptosis requires NF-kappaB inhibition and the mitochondrial permeability transition in human hepatoma cells. Hepatology 2002; 36:1498-508; PMID:12447876 [DOI] [PubMed] [Google Scholar]
- 17.Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, Lee D, von Goetz M, Yee SF, Totpal K, Huw L, Katta V, Cavet G, Hymowitz SG, Amler L, Ashkenazi A. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med 2007; 13:1070-7; PMID:17767167; http://dx.doi.org/ 10.1038/nm1627 [DOI] [PubMed] [Google Scholar]
- 18.Jin Z, McDonald ER, Dicker DT, El-Deiry WS. Deficient tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor transport to the cell surface in human colon cancer cells selected for resistance to TRAIL-induced apoptosis. J Biol Chem 2004; 279:35829-39; PMID:15155747; http://dx.doi.org/ 10.1074/jbc.M405538200 [DOI] [PubMed] [Google Scholar]
- 19.Wu F, Hu Y, Long J, Zhou YJ, Zhong YH, Liao ZK, Liu SQ, Zhou FX, Zhou YF, Xie CH. Cytotoxicity and radiosensitization effect of TRA-8 on radioresistant human larynx squamous carcinoma cells. Oncol Rep 2009; 21:461-5; PMID:19148523; http://dx.doi.org/ 10.3892/or_00000347 [DOI] [PubMed] [Google Scholar]
- 20.Hu Y, Ouyang W, Wu F, Cao CH, Wang K, Liao ZK, Zhong YH, Zhou FX, Liu SQ, Xia L, Zhou YF, Xie CH. Enhanced radiosensitivity of SW480 cells via TRAIL up-regulation mediated by Egr-1 promoter. Oncol Rep 2009; 22:765-71; PMID:19724854 [DOI] [PubMed] [Google Scholar]
- 21.Voelkel-Johnson C, Hannun YA, El-Zawahry A. Resistance to TRAIL is associated with defects in ceramide signaling that can be overcome by exogenous C6-ceramide without requiring downregulation of cellular FLICE inhibitory protein. Mol Cancer Ther 2005; 4:1320-7; PMID:16170023; http://dx.doi.org/ 10.1158/1535-7163.MCT-05-0086 [DOI] [PubMed] [Google Scholar]
- 22.Ipatova OM, Torkhovskaya TI, Zakharova TS, Khalilov EM. Sphingolipids and cell signaling: involvement in apoptosis and atherogenesis. Biochemistry (Mosc) 2006; 71:713-22; PMID:16903825; http://dx.doi.org/ 10.1134/S0006297906070030 [DOI] [PubMed] [Google Scholar]
- 23.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol 2008; 9:139-50; PMID:18216770; http://dx.doi.org/ 10.1038/nrm2329 [DOI] [PubMed] [Google Scholar]
- 24.Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind S, Spiegel S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 1996; 381:800-3; PMID:8657285; http://dx.doi.org/ 10.1038/381800a0 [DOI] [PubMed] [Google Scholar]
- 25.Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol 2003; 4:397-407; PMID:12728273; http://dx.doi.org/ 10.1038/nrm1103 [DOI] [PubMed] [Google Scholar]
- 26.Spiegel S, Milstien S. Sphingosine 1-phosphate, a key cell signaling molecule. J Biol Chem 2002; 277:25851-4; PMID:12011102; http://dx.doi.org/ 10.1074/jbc.R200007200 [DOI] [PubMed] [Google Scholar]
- 27.Kohama T, Olivera A, Edsall L, Nagiec MM, Dickson R, Spiegel S. Molecular cloning and functional characterization of murine sphingosine kinase. J Biol Chem 1998; 273:23722-8; PMID:9726979; http://dx.doi.org/ 10.1074/jbc.273.37.23722 [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Sugiura M, Nava VE, Edsall LC, Kono K, Poulton S, Milstien S, Kohama T, Spiegel S. Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J Biol Chem 2000; 275:19513-20; PMID:10751414; http://dx.doi.org/ 10.1074/jbc.M002759200 [DOI] [PubMed] [Google Scholar]
- 29.Gault CR, Obeid LM. Still benched on its way to the bedside: sphingosine kinase 1 as an emerging target in cancer chemotherapy. Crit Rev Biochem Mol Biol 2011; 46:342-51; PMID:21787121; http://dx.doi.org/ 10.3109/10409238.2011.597737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song L, Xiong H, Li J, Liao W, Wang L, Wu J, Li M. Sphingosine kinase-1 enhances resistance to apoptosis through activation of PI3K/Akt/NF-kappaB pathway in human non-small cell lung cancer. Clin Cancer Res 2011; 17:1839-49; PMID:21325072; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-0720 [DOI] [PubMed] [Google Scholar]
- 31.Wang Q, Li J, Li G, Li Y, Xu C, Li M, Xu G, Fu S. Prognostic significance of sphingosine kinase 2 expression in non-small cell lung cancer. Tumour Biol 2014; 35:363-8; PMID:23918304; http://dx.doi.org/ 10.1007/s13277-013-1051-1 [DOI] [PubMed] [Google Scholar]
- 32.Antoon JW, White MD, Slaughter EM, Driver JL, Khalili HS, Elliott S, Smith CD, Burow ME, Beckman BS. Targeting NFkB mediated breast cancer chemoresistance through selective inhibition of sphingosine kinase-2. Cancer Biol Ther 2011; 11:678-89; PMID:21307639; http://dx.doi.org/ 10.4161/cbt.11.7.14903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.French KJ, Zhuang Y, Maines LW, Gao P, Wang W, Beljanski V, Upson JJ, Green CL, Keller SN, Smith CD. Pharmacology and antitumor activity of ABC294640, a selective inhibitor of sphingosine kinase-2. J Pharmacol Exp Ther 2010; 333:129-39; PMID:20061445; http://dx.doi.org/ 10.1124/jpet.109.163444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson KR, Johnson KY, Crellin HG, Ogretmen B, Boylan AM, Harley RA, Obeid LM. Immunohistochemical distribution of sphingosine kinase 1 in normal and tumor lung tissue. J Histochem Cytochem 2005; 53:1159-66; PMID:15923363; http://dx.doi.org/ 10.1369/jhc.4A6606.2005 [DOI] [PubMed] [Google Scholar]
- 35.Li J, Guan HY, Gong LY, Song LB, Zhang N, Wu J, Yuan J, Zheng YJ, Huang ZS, Li M. Clinical significance of sphingosine kinase-1 expression in human astrocytomas progression and overall patient survival. Clin Cancer Res 2008; 14:6996-7003; PMID:18980995; http://dx.doi.org/ 10.1158/1078-0432.CCR-08-0754 [DOI] [PubMed] [Google Scholar]
- 36.Taha TA, Hannun YA, Obeid LM. Sphingosine kinase: biochemical and cellular regulation and role in disease. J Biochem Mol Biol 2006; 39:113-31; PMID:16584625; http://dx.doi.org/ 10.5483/BMBRep.2006.39.2.113 [DOI] [PubMed] [Google Scholar]
- 37.Ozoren N, Fisher MJ, Kim K, Liu CX, Genin A, Shifman Y, Dicker DT, Spinner NB, Lisitsyn NA, El-Deiry WS. Homozygous deletion of the death receptor DR4 gene in a nasopharyngeal cancer cell line is associated with TRAIL resistance. Int J Oncol 2000; 16:917-25; PMID:10762627 [DOI] [PubMed] [Google Scholar]
- 38.Eggert A, Grotzer MA, Zuzak TJ, Wiewrodt BR, Ho R, Ikegaki N, Brodeur GM. Resistance to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in neuroblastoma cells correlates with a loss of caspase-8 expression. Cancer Res 2001; 61:1314-9; PMID:11245427 [PubMed] [Google Scholar]
- 39.Abdulghani J, El-Deiry WS. TRAIL receptor signaling and therapeutics., 2010:1091-108 [DOI] [PubMed] [Google Scholar]
- 40.Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 1997; 277:815-8; PMID:9242610; http://dx.doi.org/ 10.1126/science.277.5327.815 [DOI] [PubMed] [Google Scholar]
- 41.Dimberg LY, Anderson CK, Camidge R, Behbakht K, Thorburn A, Ford HL. On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene 2013; 32:1341-50; PMID:22580613; http://dx.doi.org/ 10.1038/onc.2012.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorli SC, Colie S, Albinet V, Dubrac A, Touriol C, Guilbaud N, Bedia C, Fabrias G, Casas J, Segui B, Levade T, Andrieu-Abadie N. The nonlysosomal β-glucosidase GBA2 promotes endoplasmic reticulum stress and impairs tumorigenicity of human melanoma cells. Faseb J 2013; 27:489-98; PMID:23073830; http://dx.doi.org/ 10.1096/fj.12-215152 [DOI] [PubMed] [Google Scholar]
- 43.Liu YY, Han TY, Giuliano AE, Cabot MC. Ceramide glycosylation potentiates cellular multidrug resistance. Faseb J 2001; 15:719-30; PMID:11259390; http://dx.doi.org/ 10.1096/fj.00-0223com [DOI] [PubMed] [Google Scholar]
- 44.Pchejetski D, Golzio M, Bonhoure E, Calvet C, Doumerc N, Garcia V, Mazerolles C, Rischmann P, Teissie J, Malavaud B, Cuvillier O. Sphingosine kinase-1 as a chemotherapy sensor in prostate adenocarcinoma cell and mouse models. Cancer Res 2005; 65:11667-75; PMID:16357178; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-2702 [DOI] [PubMed] [Google Scholar]
- 45.Ozoren N, El-Deiry WS., Defining characteristics of Types I and II apoptotic cells in response to TRAIL. Neoplasia 2002. Four(6): p. 551-7; PMID:12407450; http://dx.doi.org/ 10.1038/sj.neo.7900270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White MD, Chan L, Antoon JW, Beckman BS. Targeting ovarian cancer and chemoresistance through selective inhibition of sphingosine kinase-2 with ABC294640. Anticancer Res 2013; 33:3573-9; PMID:24023282 [PubMed] [Google Scholar]
- 47.Gao P, Peterson YK, Smith RA, Smith CD. Characterization of isoenzyme-selective inhibitors of human sphingosine kinases. PLoS One 2012; 7:e44543; PMID:22970244; http://dx.doi.org/ 10.1371/journal.pone.0044543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henson ES, Gibson EM, Villanueva J, Bristow NA, Haney N, Gibson SB. Increased expression of Mcl-1 is responsible for the blockage of TRAIL-induced apoptosis mediated by EGF/ErbB1 signaling pathway. J Cell Biochem 2003; 89:1177-92; PMID:12898516; http://dx.doi.org/ 10.1002/jcb.10597 [DOI] [PubMed] [Google Scholar]
- 49.Kreuz S, Siegmund D, Scheurich P, Wajant H. NF-kappaB inducers upregulate cFLIP, a cycloheximide-sensitive inhibitor of death receptor signaling. Mol Cell Biol 2001; 21:3964-73; PMID:11359904; http://dx.doi.org/ 10.1128/MCB.21.12.3964-3973.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azijli K, Yuvaraj S, Peppelenbosch MP, Wurdinger T, Dekker H, Joore J, van Dijk E, Quax WJ, Peters GJ, de Jong S, Kruyt FA. Kinome profiling of non-canonical TRAIL signaling reveals RIP1-Src-STAT3-dependent invasion in resistant non-small cell lung cancer cells. J Cell Sci 2012; 125:4651-61; PMID:22797920; http://dx.doi.org/ 10.1242/jcs.109587 [DOI] [PubMed] [Google Scholar]
- 51.Stegehuis JH, de Wilt LH, de Vries EG, Groen HJ, de Jong S, Kruyt FA. TRAIL receptor targeting therapies for non-small cell lung cancer: current status and perspectives. Drug Resist Updat 2010; 13:2-15; PMID:20036602; http://dx.doi.org/ 10.1016/j.drup.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 52.Hellwig CT, Rehm M. TRAIL signaling and synergy mechanisms used in TRAIL-based combination therapies. Mol Cancer Ther 2012; 11:3-13; PMID:22234808; http://dx.doi.org/ 10.1158/1535-7163.MCT-11-0434 [DOI] [PubMed] [Google Scholar]
- 53.Jin Z, McDonald ER, Dicker DT, El-Deiry WS. Deficient tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor transport to the cell surface in human colon cancer cells selected for resistance to TRAIL-induced apoptosis. J Biol Chem 2004; 279:35829-39; PMID:15155747; http://dx.doi.org/ 10.1074/jbc.M405538200 [DOI] [PubMed] [Google Scholar]
- 54.Beljanski V, Knaak C, Smith CD. A novel sphingosine kinase inhibitor induces autophagy in tumor cells. J Pharmacol Exp Ther 2010; 333:454-64; PMID:20179157; http://dx.doi.org/ 10.1124/jpet.109.163337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.